Abstract
Background
Melioidosis is gaining recognition as an emerging infectious disease with diverse clinical manifestations and high-case fatality rates worldwide. However, the molecular epidemiology of the disease outside the endemic regions such as northeast part of Thailand and northern Australia remains unclear.
Methodology/Principal findings
Clinical data and B. pseudomallei isolates obtained from 199 culture-confirmed cases of melioidosis diagnosed during 2006–2016 in South India were used to elucidate the host and pathogen specific variable virulence determinants associated with clinical presentations and disease outcome. Further, we determined the temporal variations and the influence of ecological factors on B.pseudomallei Lipopolysaccharide (LPS) genotypes causing infections. Severe forms of the disease were observed amongst 169 (85%) patients. Renal dysfunction and infection due to B.pseudomallei harboring BimABm variant had significant associations with severe forms of the disease. Diabetes mellitus, septicemic melioidosis and infection due to LPSB genotype were independent risk factors for mortality. LPSB (74%) and LPSA (20.6%) were the prevalent genotypes causing infections. Both genotypes demonstrated temporal variations and had significant correlations with rainfall and humidity.
Conclusion/Significance
Our study findings suggest that the pathogen specific virulence traits under the influence of ecological factors are the key drivers for geographical variations in the molecular epidemiology of melioidosis.
Author summary
Amidst the ambiguity of its true incidence, melioidosis is gaining importance as an emerging infection in the Indian subcontinent. Variable virulence genes contributing to varied clinical presentation and the association of LPS genotypes provides key insights regarding the disease dynamics, host and pathogen specific determinants for disease presentations and outcomes among melioidosis patients in India. The study shows the divergence of Indian B.pseudomallei strains in context to LPS genotypes compared to isolates reported worldwide. The association of LPS B with mortality foreshadows the need for understanding the cascade of immune response triggered in the presence of different LPS genotypes.
Introduction
Melioidosis is a fatal infectious disease caused by soil saprophytic bacterium, Burkholderia pseudomallei. Infection occurs mostly through the inhalation or percutaneous inoculation of the bacteria from contaminated soil or surface water. The disease manifests with diverse clinical presentations ranging from mild localized infection to fulminant sepsis. B. pseudomallei being a soil saprophyte, uses horizontal gene transfers as a mechanism for its persistence both in the environment and the host. It is possible that the virulence attributes of B.pseudomallei can significantly vary under the influence of regional environmental/ecological conditions, in turn leading to the occurrence of distinct clinical manifestations. Lipopolysaccharide (LPS) of B. pseudomallei is a well-known virulence factor that confers serum resistance and helps in evading host immune defenses during the early stages of infection. In this context, LPS is gaining recognition as a potential candidate for vaccine and diagnostic assay development [1,2]. Three genotypes of LPS namely A, B and B2 were reported previously with variations in their geographic distribution and ability to induce immune responses in animal models [3]. Burkholderia intracellular motility (BimA) and filamentous hemagglutinin gene (fhaB3) were reported previously as the significant variable virulence factors based on their geographic distribution and associations with clinical presentations [4].
Amidst the ambiguity of its true incidence, melioidosis is gaining importance as an emerging infection in the Indian subcontinent [5–7]. Sporadic cases reported from different parts of the country have shown assorted clinical presentations [7–10]. Distinct/novel sequence types of B. pseudomallei using multi-locus sequence typing(MLST) were previously reported from the south-western coastal part of India [11].This geographic region might presumably be one of the endemic hotspots of melioidosis in India reporting several cases [6,7,10]. Clinical isolates of B. pseudomallei from this region were genetically diverse from the Australian and Southeast Asian isolates and the prevalent sequence type (ST 1368) lacked significant association with any particular clinical presentation of the disease [11]. With this background, the present study documents the frequencies of variable virulence factors of B. pseudomallei and their association with clinical presentations of melioidosis and the temporal variations and influence of ecological factors on commonly occurring LPS genotypes of B. pseudomallei. Moreover, the host and pathogen-specific determinants for mortality are elucidated.
Methods
Study site and population
The present study was carried out at a tertiary care hospital with 2030 inpatient capacity that caters residents of southwestern coastal parts of Karnataka, India covering nearly 150–200 km radius of geographical area. This part of the country experiences tropical climatic condition with an annual rainfall of >4000 mm during June-October. Microbiological culture confirmed cases of melioidosis over a decade (2006–2016) were included in the study.
Ethics statement
The study was approved by the Institutional Ethical Committee of Kasturba Hospital, Manipal. All the isolates were obtained from the archived collection and the identity of the patients was kept confidential.
Study isolates
Isolates from blood culture in 97 bacteremic cases with (n = 50) or without (n = 47) other organ involvement and from other sites in 102 non-bacteremic cases were included in the study. For the extraction of bacterial DNA, QIAamp DNA mini kit (Qiagen, Hilden, Germany) was used as per the manufacturer’s instructions. Before inclusion, all the study isolates were confirmed as B. pseudomallei using a species-specific PCR targeting the TTSS1 gene cluster as described previously [12].
Detection of LPS genotypes and variable virulence genes
Collectively, we aimed at detection of LPS genotypes (A, B and B2), BimA gene variants (BimABp and BimABm) and fhaB3.
Detection of LPS genotypes
Multiplex PCR assay simultaneously detected three different LPS genotypes LPS A, B, and B2 [3]. The PCR reaction was set at a final volume of 25 μl using JumpStartTaq Ready Mix (Sigma-Aldrich).Amplification was carried out in a Master cycler gradient (Eppendorf, Hamburg, Germany) with an initial denaturation at 95°C for 10 min, followed by 35 cycles of 95°C for 30sec, 59°C for 30 sec, 72°C for 30secand a final extension step of 72°C for 7 min. The oligonucleotide primers used in the present study and the expected amplicon sizes are tabulated in Table 1.
Table 1. List of oligonucleotide primers used to detect virulence genes of B. pseudomallei.
| Virulence Determinants | Oligonucleotide sequence 5’-3’ | Amplicon Size | Reference |
|---|---|---|---|
| BimABm | BimBm F—AGCGCTTCGCGCATCTAC BIMBm R-CGCGTTAAACGCCGTACTTTC |
104bp | 4 |
| BimABp | BimBp F- GGAAGCTTTGGCGTGCATAT BimBp R- CCCATGCCTTCCTCGACTAAT |
60bp | 4 |
| fhaB3 |
fhaB3 F-GACGCGGCACGTCTGATC fhaB3 R-CGCGGATAAAACTCGGATTG |
58bp | 4 |
| LPS A | wbiE_F-TCAAACCTATCCGCGTGTCGAAGT wbiE_R-TCGTCGTCAAGAAATCCCAGCCAT |
195bp | 3 |
| LPS B | BUC3396F-AATCTTTTTCTGATTCCGTCC BUC3396RACCAGAAGACAAGGAGAAAGGCCA |
93bp | 3 |
| LPS B2 | BURP840_LPSb16-F- AACCGGGTAGTTCGCGATTAC BURP840_LPSb16-R-ATACGCCGGTGTAGAACAGTA |
364bp | 3 |
Bim A detection
Both variants of Bim A, BimABm and BimABp, were detected using previously reported PCR primers [4]. PCR reaction volumes and cycling conditions for the detection of BimA genes were similar to that of the LPS genotypes, except for a change in the annealing temperature to 56°C for 1 min. Presence of BimABm and BimABp were considered when amplicons sized 104 bp and 60 bp respectively were positive on 2.5% Agarose gel stained with 0.5% ethidium bromide.
Filamentous hemagglutinin (fha) B3
fhaB3 gene was detected using 0.3 uM of each forward and reverse primers to generate a 58 bp product [4]. Amplification was carried using similar cycling conditions as mentioned above for LPS genotypes detection. However, PCR for the detection of fhaB3 was performed separately considering the similar size of the amplicons for both BimABp and fhaB3.
Clinical, epidemiological and meteorological data
Clinical and epidemiological data were documented in structured study forms. Monthly rainfall and relative humidity data for a period of six years (2010–2016) were obtained from the Indian Meteorological Department, Pune, India.
Case definitions
Microbiological culture of blood and/ or other clinically relevant specimens was the mainstay for laboratory diagnosis of melioidosis. For analysis and reporting purposes the following case definitions were used in the present study:
Bacteremic melioidosis: Patients with positive blood cultures for B. pseudomallei, with or without culture positivity of other clinically relevant specimens.
Pulmonary melioidosis: Patients with clinical and radiological evidence of pneumonia / lower respiratory infection and isolation of B. pseudomallei from respiratory specimens and/or blood.
Neurological melioidosis: Patients with clinical and radiological evidence of infection affecting brain tissue, meninges or spinal cord and isolation of B. pseudomallei from blood, cerebrospinal fluid or exudates.
Septicemic melioidosis: Patients with features of sepsis (hyper/hypo-thermia, leukocytosis, hypotension, pulse rate >90/min, respiratory rate >18/min) and isolation of B. pseudomallei from any clinical specimen.
Osteoarticular melioidosis: Patients with bone/joint infections such as osteomyelitis and septic arthritis, and isolation of B. pseudomallei from any clinical specimen.
Localized melioidosis: Patients with skin and soft tissue (or any other single-site) infections that had no bacteremia and radiological or clinical evidence, suggestive of other organ involvement and isolation of B. pseudomallei from relevant clinical specimen.
Statistical analysis
Descriptive statistical tools were used to determine the frequencies of categorical study variables. Pearson’s Chi-square test and Fisher’s exact test was used to check for the presence of any significant association of host and pathogen characteristics with individual clinical presentations. Risk factors for various clinical presentations and outcomes (in hospital mortality & Discharge Against Medical Advice) among the study population were determined using univariate analysis and step-wise multivariate logistic regression model (Backward LR) (SPSS, version 16). Time series analysis was used to decompose the trend, seasonal and residual components for the climate data and the LPS genotypes. Correlations of the climate data with the LPS genotypes was obtained using Pearson’s correlation coefficient. Generalized additive model was used to predict the LPS genotypes using the climate data. Analysis was carried out using R version 3.3.3. All values were considered significant with p≤0.05.
Results
Baseline demography and clinical data
Mean age of our patients was 47.7±15.4 years, with an age range between 7–86 years. Of the 199 patients, 187 (94%) were from Karnataka and 12 patients (8 from Kerala and 4 from Goa) were from other states on the southwestern coastal part of India (Fig 1). Majority of the patients were males (153, 76.9%) and had the disease episode during monsoon (148, 74.3%).
Fig 1. Geographical distribution of study patients and infecting LPS genotypes (Map generated using http://landsatlook.usgs.gov/.).
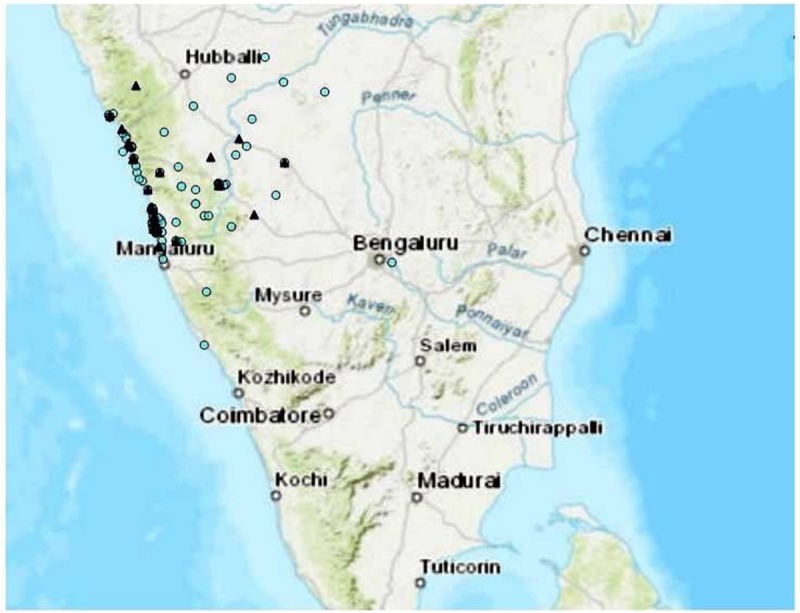
LPSB genotypes: Light Blue circle, LPSA genotypes: Blue Triangles.
A large number of patients (169, 85%) had either one or more of the severe forms of the disease, like bacteremic (97, 48.7%), pulmonary (71, 35.6%), septicemic (50, 25.1%) neurological (22, 11%) and others like osteoarticular and deep seated abscess (36, 18%). Amongst bacteremic patients, 47 (48.4%) had no other focus of infection as diagnosed clinically or radiologically. In septicemic melioidosis, 38 (76%) patients had bacteremia with or without evident foci and the rest 12 patients had osteoarticular or pulmonary forms of the disease. Localized form of the disease was observed amongst 30 (15%) patients.
Association of host factors with clinical presentations
Diabetes mellitus (DM) (124, 62.3%) and renal dysfunction (27, 13.5%) were the common co-morbid illnesses. Three patients had malignancy, one had thalassemia, and none had HIV infection. Using univariate analysis, we observed that patients with renal dysfunction had 8.75 (Crude OR:8.75; 95% CI: 3.60–21.25; p<0.001) and 3.52 (Crude OR: 3.52; 95% CI: 1.41–8.77; p = 0.005) times more odds for developing septicemic and bacteremic forms of the disease respectively(Table 2).
Table 2. Association of demographical, seasonal and premorbid illnesses with clinical characteristics in study population.
| Variables (N = 199) | Bacteremia (n = 97) |
Pulmonary (n = 71) |
Neurological (n = 22) | Sepsis (n = 50) | Localized (n = 30) |
OA & DOA# (n = 36) |
|---|---|---|---|---|---|---|
|
Age groups in yrs <18 (14) 19–50 (99) >51 (86) p-value OR (95%CI)* |
5 (35.7) 43 (43.4) 49 (57) 0.111 1.72(0.96–3.09) |
5 (35.7) 35 (35.4) 31 (36) 0.995 0.97(0.53–1.77) |
0 14 (14.1) 8 (9.3) 0.227 1.60(0.63–4.03) |
2 (14.3) 24 (24.2) 24 (27.2) 0.530 1.21(0.62–2.33) |
4 (28.6) 17 (17.2) 9 (10.5) 0.153 1.77(0.74–4.21) |
4 (28.6) 18 (18.2) 14 (16.3) 0.541 1.14(0.53–2.46) |
|
Gender Male (153) Female (46) p-value OR (95% CI)* |
73 (47.7) 24 (52.2) 0.596 0.83 (0.43–1.61) |
53 (34.6) 18 (39.1) 0.577 1.21(0.61–2.39) |
17 (11.1) 5 (10.9) 0.963 0.97(0.33–2.80) |
34 (22.2) 16 (34.8) 0.085 0.53(0.26–1.09) |
22 (14.4) 8 (17.4) 0.617 1.25(0.51–3.04) |
32 (20.9) 4 (8.7) 0.059 0.36(0.12–1.07) |
|
Season Monsoon (148) Dry (51) p-value OR (95% CI)* |
78 (52.7) 19 (37.3) 0.057 1.87(0.97–3.60) |
56 (37.8) 15 (29.4) 0.279 0.68(0.34–1.36) |
17 (11.5) 5 (9.8) 0.741 0.83(0.29–2.39) |
41 (27.7) 9 (17.6) 0.153 1.78(0.80–3.99) |
22 (14.9) 8 (15.7) 0.888 1.06(0.44–2.56) |
23 (15.5) 13 (25.5) 0.111 1.85(0.86–4.01) |
|
Diabetes mellitus Yes (124) No (75) p-value OR (95% CI)* |
61 (49.2) 36 (48) 0.870 1.04(0.59–1.86) |
49 (39.5) 22 (29.3) 0.146 0.63(0.34–1.17) |
14 (11.3) 8 (10.7) 0.892 0.93(0.37–2.35) |
36 (29) 14 (18.7) 0.102 1.78(0.88–3.58) |
18 (14.5) 12 (16) 0.777 1.12(0.57–1.50) |
26 (21) 10 (13.3) 0.175 0.58(0.26–1.28) |
|
Renal dysfunction Yes (27) No (172) p-value OR (95% CI)* |
20 (74.1) 77 (44.8) 0.005 3.52(1.41–8.77) |
15 (55.6) 56 (32.6) 0.02 0.38(0.17–0.88) |
0 22 (100) 0.049 0.84(0.79–0.90) |
18 (66.7) 32 (18.6) <0.001 8.75(3.60–21.25) |
3 (11.1) 27 (15.7) 0.773 1.49(0.41–5.29) |
5 (18.5) 31 (18) 1.00 0.96(0.34–2.75) |
#Osteoarticular and Deep Organ Abscess
* Crudes Odds Ratio is reported for all the variables
p value was calculated using Chi square or Fisher’s exact test.
Frequencies of individual virulence factors and their association with clinical presentations
Majority of our study isolates belonged to LPS B (n = 147, 73.8%) followed by A (n = 41, 20.6%) and B2 (n = 11, 5.5%) genotypes. Amongst the variants of BimA gene, BimABp and BimABm were observed among 190 (95.4%) and 9 (4.5%) of the isolates respectively. Majority of the isolates were positive for fhaB3(190; 95.4%) and 181 (90.4%) isolates harbored both BimABp and fhaB3genes. None of the three LPS genotypes had a significant association with clinical forms of the disease in our study population, as it was observed withBimA gene variants with neurological form of the disease(Crude OR: 12.72; 95% CI: 3.11–51.89; p>0.001)(Table 3).
Table 3. Association of lipopolysaccharide genotypes and variable virulence genes of B. pseudomallei isolates with clinical presentations in our study.
| Variables (N = 199) | Bacteremia (n = 97) |
Pulmonary (n = 71) |
Neurological (n = 22) | Sepsis (n = 50) | Localized (n = 30) |
OA & DOA# (n = 36) |
|---|---|---|---|---|---|---|
| LPS A Yes (41) No (158) p-value OR (95% CI) |
20 (48.8) 77 (47.7) 0.996 0.99(0.50–1.98) |
17 (41.5) 54 (34.2) 0.386 1.36(0.67–2.75) |
5 (12.2) 17 (10.8) 0.794 1.15(0.39–3.32) |
11 (26.8) 39 (24.6) 0.492 0.76(0.35–1.64) |
5 (12.2) 25 (15.8) 0.563 0.73(0.26–2.06) |
9 (22) 27 (17.1) 0.471 1.36(0.58–3.18) |
| LPS B Yes (147) No (52) p-value OR (95% CI) |
72 (49) 25 (48.1) 0.911 0.96(0.51–1.81) |
52 (35.4) 19 (36.5) 0.880 0.95(0.49–1.83) |
17 (11.6) 5 (9.6) 0.700 1.22(0.43–3.51) |
36 (24.5) 14 (26.9) 0.728 1.13(0.55–2.33) |
24 (16.3) 6 (11.5) 0.407 1.49(0.57–3.89) |
26 (17.7) 10 (19.2) 0.804 0.90(0.40–2.02) |
| LPS B2 Yes (11) No (188) p-value OR (95% CI) |
6 (54.5) 91 (48.4) 0.692 0.78(0.23–2.65) |
2 (18.2) 69 (36.7) 0.334 0.38(0.08–1.82) |
0 22 (11.7) 0.615 0.93(0.90–0.97) |
3 (27.3) 47 (25) 1.00 0.88(0.22–3.48) |
1 (9.1) 29 (15.4) 1.00 0.54(0.06–4.44) |
1 (9.1) 35 (18.6) 0.693 0.43(0.05–3.52) |
|
BimA variants BimABp(190) BimABm (9) p-value OR (95% CI) |
93 (48.9) 4 (44.4) 1.00 0.83(0.21–3.20) |
69 (36.3) 2 (22.2) 0. 388 1.99(0.40–9.87) |
17 (8.9) 5 (55.6) 0.001 12.72(3.11–51.89) |
48 (25.3) 2 (22.2) 1.00 0.84(0.17–4.20) |
27 (14.2) 3 (33.3) 0.138 0.33(0.07–1.40) |
35 (18.4) 1 (11.1) 1.00 1.80(0.21–14.91) |
|
fhaB3 Yes (190) No (9) p-value OR (95% CI) |
94 (49.5) 3 (33.3) 0.344 0.51(0.12–2.10) |
68 (35.8) 3 (33.3) 1.00 1.15(0.27–4.59) |
21 (11.1) 1 (11.1) 1.00 0.99(0.11–8.34) |
47 (24.7) 3 (33.3) 0.694 1.52(0.36–6.32) |
30 (15.8) 0 0.360 0.94(0.91–0.98) |
33 (17.4) 3 (33.3) 0.209 0.42(0.10–1.76) |
#Osteoarticular and Deep Organ Abscess.Crudes Odds Ratio is reported for all the variables
p value was calculated using Chi square or Fisher exact test.
Spatial and temporal variations of LPS genotypes
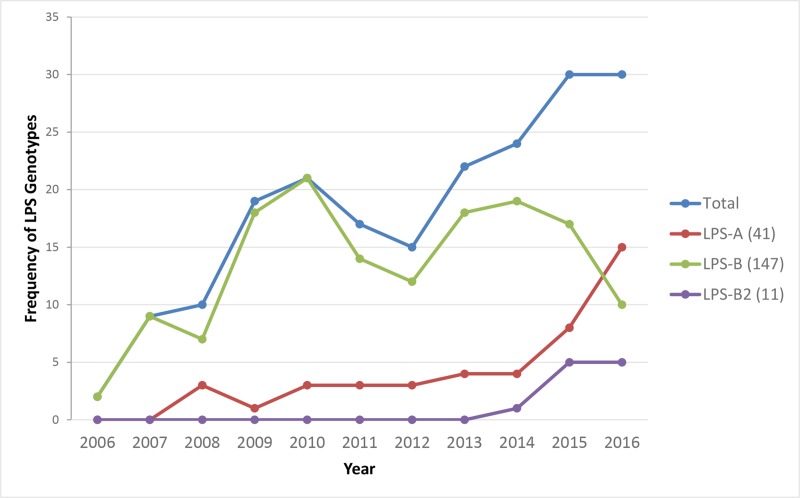
Majority (73.8%) of the B. pseudomallei clinical isolates including all the 12 isolates from patients in the adjacent states belonged to the LPSB genotype (Fig 1). The prevalence of LPS A and LPS B genotypes were consistent throughout the study period (2006–2016), and LPS B2 genotype was observed only during the last three years (2014–2016). We noticed a steady decline of LPSB during the years 2014–2016 (79% in 2014, 56.6% in 2015 and 50% in 2016) in our settings. At the same time, there was a steady increase ofLPSA (16.6%, 26.6% and 33.3%) and B2 (4.1%, 16.6% and 16.6%) genotypes during the same duration (Fig 2).
Fig 2. Year-wise distribution (2006–2016) of B.pseudomallei lipopolysaccharide genotypes causing infections in our settings.
Influence of monthly rainfall and humidity on lipopolysaccharide diversity
Monthly rainfall and humidity data for the years 2010–2016 was plotted against time and decomposed data for trend, seasonality and residual component were compared with LPSA and B genotypes. LPSA genotype showed a reversal peak for trend and seasonal components of rainfall, whereas rainfall had a significant increasing effect on LPSB genotype. There was a rise in LPSA genotypes observed over time, as compared to the LPSB genotypes. On omitting the seasonal trends, the residual component showed a positive effect on LPSB (p<0.001). The trend (p = 0.04) and seasonality(p = 0.01) for rainfall also showed a positive effect on LPSB genotype, whereas seasonality only had a positive correlation (p<0.001) for LPSA (Figs 3 & 4). Comparing the patterns of LPS genotypes with humidity, a positive correlation was observed between the seasonal component of humidity with LPSA (p<0.001) and LPSB (p = 0.003). A lag period was observed in the peaks of both LPSA and B genotypes compared to trend and seasonal component of rainfall.
Fig 3.
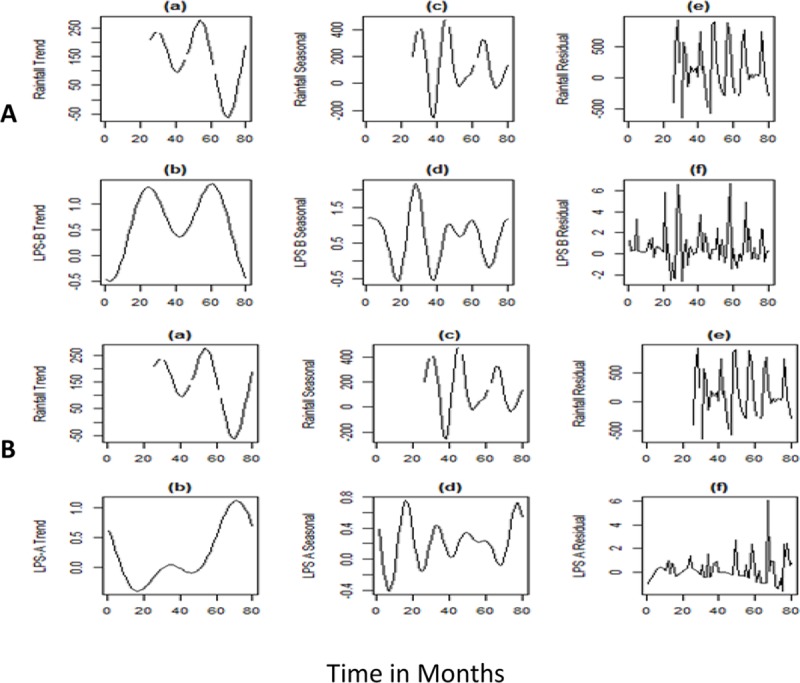
Fig A) shows reversal peaks for the trend and seasonal components of rainfall and LPS-A. B) shows an increase in LPS-B genotype with trend and seasonality. There is a lag period between the peak rainfall and the LPS-A and LPS-B genotypes.
Fig 4.
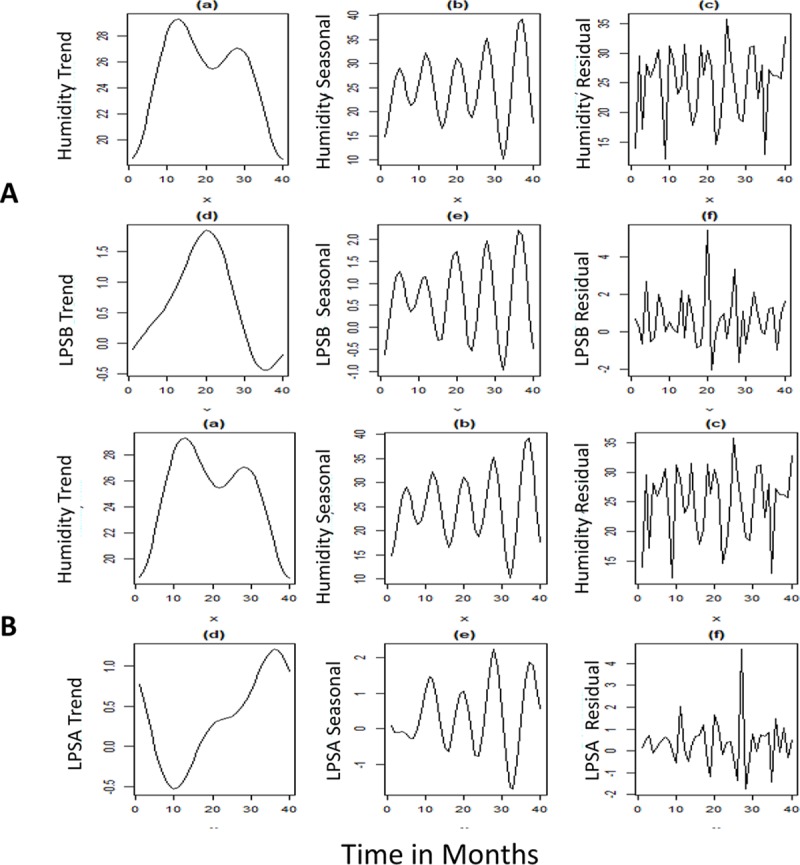
Fig A) shows reversal peaks for the trend component of humidity and LPS-A. B) shows an increase in LPS-B genotype with trend, seasonality and the residual component. There is a lag period between the peak humidity and LPS-B genotypes.
Treatment and outcomes
Among the 199 cases, 173 (87%) patients received melioidosis specific therapy. Of the 26 septicemic patients who did not receive pathogen specific therapy, 17 and 9 cases were with and without bacteremia respectively. Mortality due to melioidosis was observed among 51 (25.1%) patients. None of the patients with localized form of the disease had adverse clinical outcomes. Among the 50 patients with septicemic melioidosis, 23 (46%) succumbed to death. Case fatality rates were 14.5% (n = 25) among patients who received pathogen-specific therapy in comparison to 100% (n = 26) among those who did not receive the specific therapy.
Host and pathogen specific determinants for clinical presentations and mortality
Renal dysfunction was an independent risk factor (after considering all the host and pathogen characteristics) for both bacteremic (Adjusted OR: 3.52 (1.41–8.87), p = 0.007) and septicemic (Adjusted OR: 9.70 (3.88–24.22), p<0.001) forms of the disease (Tables 2 & 3). Infection due to B. pseudomallei having BimABm variant (Adj OR: 14.41 (3.16–65.58), p = 0.001) was an independent risk factor for neurological melioidosis in our study population. DM(26/51; 51%)[Adjusted OR: 2.37 (95%CI: 1.19–4.74), p = 0.014], septicemic melioidosis(32/51; 63%)[Adjusted OR: 2.74 (95%CI: 1.31–5.85), p = 0.007] and infection due to B. pseudomallei LPSB genotype (46/51; 90.1%) [Adjusted OR: 4.47 (95%CI: 1.16–12.20), p = 0.003] were independent risk factors for mortality in our study population.
Discussion
B. pseudomallei, the etiological agent of melioidosis, demands no further negligence in view of its increasing geographical distribution, possession of numerous virulence traits and intrinsic resistance mechanisms to several antimicrobial agents. Severity of the infection and disease outcomes among patients significantly depend on the underlying host-factors, time for diagnosis/detection and befitting medical management. Few hostfactors such as DM, renal dysfunction and chronic alcoholism are well associated with poor disease outcomes among patients with melioidosis. B.pseudomallei possesses numerous proteins that play a pivotal role in the pathogenesis of the disease [4].Some genes that encode these proteins (conferring virulence) are known to be ubiquitously present among all the B. pseudomallei isolates [4].More recently, a study on the global evolution of B. pseudomallei reported geographically distinct genes/variants, conferring virulence among Australasian and Southeast Asian isolates [13]. However, there is a visible scant of whole genome sequencing data and the virulence attributes of B. pseudomallei isolates from regions outside northern Australia and northeast Thailand. Numerous studies have reported the influence of ecological factors on B. pseudomallei positivity in environmental niches [14, 15]. However, it is currently unknown if there is any influence of the ecological factors on the variable virulence genes of B. pseudomallei and whether these variations in the virulence gene profiles can lead to distinct clinical presentations and outcomes. Given this context, we report here the correlation of ecological factors in a geographical locality on infecting LPS genotypes of B. pseudomallei amongst patients.
Lipopolysaccharide of B. pseudomallei is an important virulence factor that facilitates the evasion of human immune responses during the early stages of infection. Monoclonal antibodies against the LPS of B. pseudomallei were found to reduce the severity of disease in animal models, thus implying the role of LPS as a potential vaccine candidate [1].Most intriguing finding from our study is that the majority (74%) of our patients were infected by the LPSB genotype of B. pseudomallei. This observation is in contrast with the findings from Thailand (2.3%) and Australia (13.8%), where LPSA was reported to be the prevalent infecting genotype [4]. Immunological responses and disease outcomes among animal models administered with LPS A and B types of B. pseudomallei were reported previously. While LPSA is known to confer serum resistance and grow in the presence of 10–30% of normal human serum [16], evidence from recent experimental and animal model studies suggest that LPSB is a more potent inducer of the pro-inflammatory cytokines and septic-shock [17]. In our previous study ST 1368 was the most common sequence type observed among 32 B.pseudomallei isolates [11]. Out of 14 ST 1368, 12 (85.7%) were LPSB,2 (14.2%) were LPSA and none were LPSB2. This finding suggests that the LPS genotypes can vary among isolates belonging to the same sequence type and thus making it difficult to ‘brand’ a particular ST as either a more or a less virulent one. In the present study, we did not observe a significant association of any of the three genotypes (A, B and B2) with any particular clinical presentation. We observed that infection due to LPSB genotype was an independent risk factor for mortality among our study population. However, we foresee the need for further validating this finding amongst patient populations from other geographic locations.
Expression of Burkholderia intracellular motility (BimA) protein is crucial in the pathogenesis of the disease. Among the two variants of Bim A known, BimABp was the only variant reported among isolates from Thailand and other South Asian countries. On the contrary, isolates from Australia were reported to have both BimABp and BimABm variants [4]. Among our study isolates BimABp variants were more commonly observed, but with no association with any particular clinical form of the disease. Presence of BimABm was also observed in few (n = 9) of our isolates and had a significant association with neurological presentations. Similar association of BimABm variant with neurological melioidosis was reported in Australian patients and more recently in a study using mice model [4, 18]. Filamentous hemagglutinin (FHA) is a surface protein of B. pseudomallei involved in adhesion to the host epithelial cells and formation of multinucleated giant cells [19].fhaB3 is one of the three variable genes responsible for encoding the FHA protein, which was reported previously among 100% and 83% of the isolates from Thailand and Australia respectively [4]. Further, presence of fhaB3 gene was reported in all the B. pseudomallei isolates obtained from Thai patients with bacteremic form of the disease and absence of fhaB3 gene was reported to have a significant association with cutaneous melioidosis among Australian patients [4]. Presence of fhaB3 gene was observed in almost 95% of our study isolates with no significant association with any form of the disease.
The epidemiology of melioidosis is characterized by environmental factors where rainfall plays a key role in transmission of the disease [20]. Majority of the cases in other endemic nations are known to occur during the monsoon when patients acquire the disease via inhalation or inoculation of the bacteria from soil and water. Occurrence of cases during the dry season, in many instances, is considered as a consequence of long latency and activation of the pathogen from latent foci [21]. In the present study, we observed that the LPSA genotypes had a reverse correlation with rainfall. This finding suggests the possibility of presence of LPSA genotype in dry environmental conditions, unlike the LPSB genotypes, which had a positive correlation with rainfall and humidity. However, we did not observe a significant increase in the occurrence of infections due to the LPSA genotypes during the dry season to support the assumption. Further analyzing the data, a lag period was observed between the occurrence of the cases and rainfall. The observed lag period could not be determined due to lack of enough data points and unavailability of weekly rainfall data, which remains as one of the limitations of our study. Temperature did not show any correlation with the infecting genotypes in our study, which can be attributed to the absence of striking variations in temperature through the year across the western coastal part of the country. Upon dismissing the seasonal trends, LPSB showed a positive correlation with the residual component, suggesting an influence of other environmental factors along with rainfall and humidity which needs further investigations.
Among the co-morbid illnesses observed in the present study population, renal dysfunction was found to be an independent risk factor for septicemic and bacteremic forms of the disease. DM was found to be an independent risk factor for mortality due to melioidosis in our settings, as reported in patients from Thailand and northern parts of Australia [22,23]. Strong association of DM with mortality was not surprising since 62% of the 169 patients with severe form of the disease were diabetic in our study cohort. However, DM did not have any significant association with any one particular form of the disease. Considering the high prevalence (16%) of DM amongst adult population residing in our settings [24], we foresee the need of more focused clinical studies to understand the disease outcome amongst patients with controlled and uncontrolled DM.
In our study cohort, though not statistically significant, we observed localized melioidosis amongst a higher proportion of patients with age <18 years (28.6%) in comparison with those with age >51 years (10.5%). Skin/soft tissue infections were the common clinical presentations among cases of localized melioidosis in our settings. These infections are more likely to occur in children due to their exposure to the bacteria while playing in water lodged fields during monsoon season. Higher frequency of localized, non-bacteremic cases were previously reported among Australian patients belonging to age group of <16 years [25]. Increased occurrence of localized form of the disease among patients of < 18 years of age can also be attributed to the lack of other predisposing factors responsible for the dissemination.
Put together, the present study reports few important host and pathogen-specific virulence determinants that have significant associations with clinical presentations and disease outcomes among Indian patients infected with B.pseudomallei. These findings can help in identifying high-risk cohort of patients for future studies aiming to understand the molecular pathogenesis mechanisms of the disease using integrated omics based approaches.
Acknowledgments
The authors would like to acknowledge the contributions of the clinicians of Kasturba Hospital, Manipal in treating the patients with melioidosis. The authors also acknowledge Ms. Snigdha Reddy, Postgraduate, Department of Microbiology, Kasturba Medical College, Manipal for providing assistance in laboratory work. We would also like to acknowledge the National Data Center, Indian Metrological department Pune, India for providing the seasonal data for rainfall, humidity and temperature.
Data Availability
All relevant data are within the manuscript.
Funding Statement
The authors received no specific funding for this work.
References
- 1.AuCoin DP, Reed DE, Marlenee NL, Bowen RA, Thorkildson P, Judy BM et al. Polysaccharide Specific Monoclonal Antibodies Provide Passive Protection against Intranasal Challenge with Burkholderia pseudomallei. PLoS ONE 2012; 7(4): e35386 10.1371/journal.pone.0035386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chantratita N, Tandhavanant S, Myers ND, Seal S, Arayawichanont A, Kliangsa-ad Aet al. Survey of Innate Immune Responses to Burkholderia pseudomallei in Human Blood Identifies a Central Role for Lipopolysaccharide. PLoS ONE 2013; 8(11): e81617 10.1371/journal.pone.0081617 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tuanyok A, Stone JK, Mayo M, Gruendike J, Georgia S et al. The Genetic and Molecular Basis of O-Antigenic Diversity in Burkholderia pseudomallei Lipopolysaccharide. PLoSNegl Trop Dis 2012;6(1): e1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sarovich DS, Price EP, Webb JR, Voutsinos MY, Tuanyok A et al. Variable Virulence Factors in Burkholderia pseudomallei (Melioidosis) Associated with Human Disease. PLoS ONE 2014; 9(3): e91682 10.1371/journal.pone.0091682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Limmathurotsakul D, Golding N, Dance DA,Pigott DM, Moyes CL, Rolim DB et al. Predicted global distribution of Burkholderia pseudomallei and burden of melioidosis. NatMicrobiol. 2016;1(1). [DOI] [PubMed] [Google Scholar]
- 6.Mukhopadhyay C, Chawla K, Sushma Krishna, Rao SP, Bairy I. Emergence of Burkholderia pseudomallei and pandrug-resistant non-fermenters from southern Karnataka, India.Trans R Soc Trop Med Hyg., Volume 102, Issue Supplement_1, 1 December 2008, Pages S12–S17. [DOI] [PubMed] [Google Scholar]
- 7.Vidyalakshmi K, Lipika S, Vishal S, Chakrapani M. Emerging clinico-epidemiological trends in melioidosis: analysis of 95 cases from western coastal India. Int J Infect Dis. 2012;16(7): e491–7. 10.1016/j.ijid.2012.02.012 [DOI] [PubMed] [Google Scholar]
- 8.Gopalakrishnan R, Sureshkumar D, Thirunarayan M, Ramasubramanian V. Melioidosis: an emerging infection in Indian J Assoc Physicians India. 2013;61(9):24–6. [PubMed] [Google Scholar]
- 9.Viswaroop BS, Balaji V, Mathai E, Kekre NS. Melioidosis presenting as genitourinary infection in two men with diabetes. J Postgrad Med.2007; 4;53(2):108 [DOI] [PubMed] [Google Scholar]
- 10.Mukhopadhyay C, Shaw T, Varghese G, Dance D. Melioidosis in South Asia (India, Nepal, Pakistan, Bhutan and Afghanistan). Trop Med Infect Dis. 2018;3(2):51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tellapragada C, Kamthan A, Shaw T, Vandana KE, Kumar S, Bhat V, Mukhopadhyay C. Unravelling the Molecular Epidemiology and Genetic Diversity among Burkholderia pseudomallei Isolates from South India Using Multi-Locus Sequence Typing. PLos One. 2016;19;11(12): e0168331 10.1371/journal.pone.0168331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tellapragada C, Shaw T, D'Souza, Eshwara VK, Mukhopadhyay C A. Improved detection of Burkholderia pseudomallei from non-blood clinical specimens using enrichment culture and PCR: narrowing diagnostic gap in resource-constrained settings. Trop Med Int Health. 2017;22(7):866–870. 10.1111/tmi.12894 [DOI] [PubMed] [Google Scholar]
- 13.Chewapreecha C, Holden MT, Vehkala M, Välimäki N, Yang Z, Harris SRet al. Global and regional dissemination and evolution of Burkholderia pseudomallei.Nat Microbiol.2017;23; 2:16263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ngamsang R, Potisap C, Boonmee A, Lawongsa P, Chaianunporn T, Wongratanacheewin S et al. The contribution of soil physicochemical properties to the presence and genetic diversity of Burkholderia. pseudomallei. Southeast Asian J Trop Med Public Health. 2015;46(1):38–50. [PubMed] [Google Scholar]
- 15.Kaestli M, Harrington G, Mayo M, Chatfield MD, Harrington I, Hill A,et al. What drives the occurrence of the melioidosis bacterium Burkholderia pseudomallei in domestic gardens? PLoSNegl Trop Dis. 2015. 24;9(3): e0003635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.DeShazer D, Brett PJ, Woods DE. The type II O-antigen polysaccharide moiety of Burkholderia pseudomallei lipopolysaccharide is required for serum resistance and virulence. MolMicrobiol. 1998;30(5):1081–100. [DOI] [PubMed] [Google Scholar]
- 17.Norris MH,Schweizer HP, Tuanyok A. Structural diversity of Burkholderia pseudomallei lipopolysaccharides affects innate immune signaling. PLoSNegl Trop Dis 2017; 11(4): e0005571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morris JL, Fane A, Sarovich DS, Price EP, Rush CM, Govan BL, et al. Increased Neurotropic Threat from Burkholderia pseudomallei Strains with a B. mallei-like Variation in the bimA Motility Gene in Australia. Emerg Infect Dis. 2017;23(5). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sim SH, Yu Y, Lin CH, Karuturi RK, Wuthiekanun V, Tuanyok A, et al. The core and accessory genomes of Burkholderia pseudomallei: implications for human melioidosis. PLoSPathog. 2008;4(10): e1000178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sim SH, Yu Y, Lin CH, Karuturi RK, Wuthiekanun V, Tuanyok A, et al. Association of melioidosis incidence with rainfall and humidity, Singapore, 2003–2012. Emerg Infect Dis 2015;21(1):159 10.3201/eid2101.140042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fisher DA, Harris PN. Melioidosis: refining management of a tropical time bomb. The Lancet. 2014; 1;383(9919): 762–4. [DOI] [PubMed] [Google Scholar]
- 22.Suputtamongkol Y, Chaowagul W, Chetchotisakd P, Lertpatanasuwun N, Intaranongpai S, Ruchutrakool T et al. Risk factors for melioidosis and bacteremic melioidosis. Clin Infect Dis. 1999;29(2):408–13. 10.1086/520223 [DOI] [PubMed] [Google Scholar]
- 23.Currie BJ, Ward L, Cheng AC. The epidemiology and clinical spectrum of melioidosis: 540 cases from the 20-year Darwin prospective study. PLoSNegl Trop Dis. 2010; 30;4(11): e900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rao CR, Kamath VG, Shetty A, Kamath A. A study on the prevalence of type 2diabetes in coastal Karnataka. Int J Diabetes DevCtries. 2010;30(2):80–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.McLeod C, Morris PS, Bauert PA, Kilburn CJ, Ward LM, Baird RW, Currie BJ. Clinical presentation and medical management of melioidosis in children: a 24-year prospective study in the Northern Territory of Australia and review of the literature. Clin Infect Dis. 2015. 1;60(1):21–6. 10.1093/cid/ciu733 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All relevant data are within the manuscript.