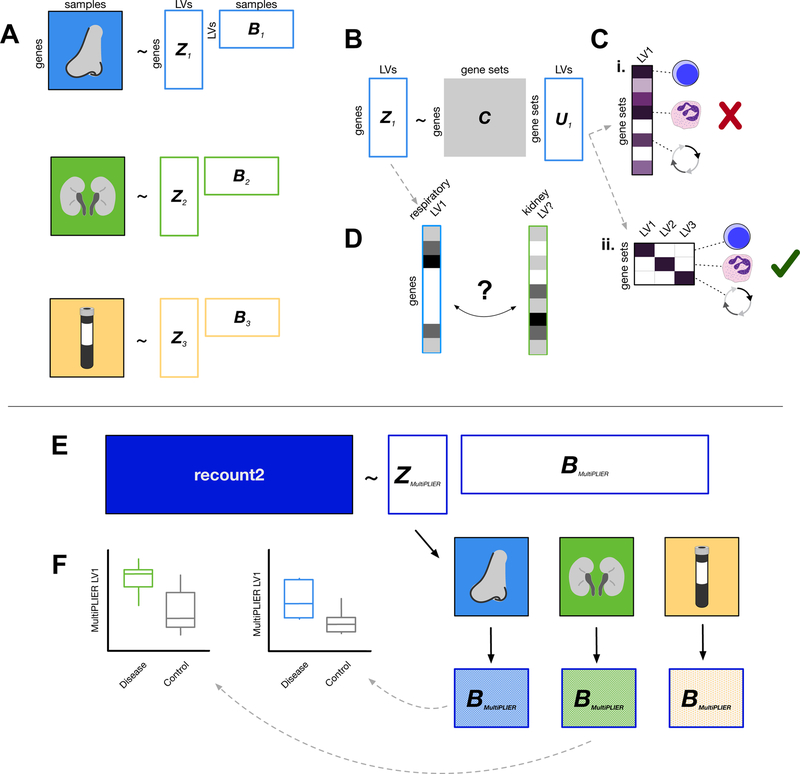
Figure 1. Overview of dataset-specific PLIER and MultiPLIER.
Boxes with solid colored fills represent inputs to the model. White boxes with colored outlines represent model output. (A) PLIER (Mao et al., 2017) automatically extracts latent variables (LVs), shown as the matrix B, and their loadings (Z). We can train PLIER model for each of three datasets from different tissues, which results in three dataset-specific latent spaces. (B) PLIER takes as input a prior information/knowledge matrix C and applies a constraint such that some of the loadings (Z) and therefore some of the latent variables capture biological signal in the form of curated pathways or cell type-specific gene sets. (C) Ideally, a latent variable will map to a single gene set or a group of highly related gene sets to allow for easy interpretation of the model. PLIER applies a penalty on U to facilitate this. Purple fill in a cell indicates a non-zero value and a darker purple indicates a higher value. We show an undesirable U matrix in the top toy example (Ci) and a favorable U matrix in the bottom toy example (Cii). (D) If models have been trained on individual datasets, we may be required to find “matching” latent variables in different dataset- or tissue-specific models using the loadings (Z) from each model. Using a metric like the Pearson correlation between loadings, we may or may not be able to find a well-correlated match between datasets. (E) The MultiPLIER approach: train a PLIER on a large collection of uniformly processed data from many different biological contexts and conditions (recount2; Collado-Torres et al., 2017)—a MultiPLIER model—and then project the individual datasets into the MultiPLIER latent space. The hatched fill indicates the sample dataset of origin. (F) Latent variables from the MultiPLIER model can be tested for differential expression between disease and controls in multiple tissues.