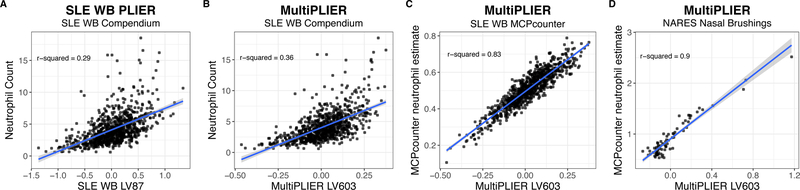
Figure 3. MultiPLIER learns a neutrophil-associated latent variable (LV) that is well-correlated with neutrophil counts or estimates in multiple tissues and disease contexts.
(A) SLE WB LV87, LV87 from the PLIER model trained on the entire SLE WB compendium, can predict neutrophil count in Banchereau, et al. dataset (Banchereau et al., 2016) despite being entirely unsupervised with respect to this task. (B) The MultiPLIER neutrophil-associated latent variable (LV603) performs slightly better than the SLE WB-specific model. Here, the SLE WB compendium is projected into the MultiPLIER latent space. Recall that the MultiPLIER model has not been exposed to the specific technical variance found in the SLE WB compendium. (C) MultiPLIER LV603 values are highly correlated from a state-of-the-art method for estimating immune infiltrate, MCPcounter (Becht et al., 2016). This suggests that the modest correlation with neutrophil count in B is the result of estimating neutrophil, a terminally differentiated cell type, counts from transcriptome data, rather than a limitation of the MultiPLIER approach. (D) MultiPLIER performance is not limited to whole blood, as it is highly correlated with the MCPcounter neutrophil estimate in the NARES dataset (Grayson et al., 2015). NARES is a nasal brushing microarray dataset that includes patients with ANCA-associated vasculitis, patients with sarcoidosis and healthy controls among other groups and was projected into the MultiPLIER latent space.