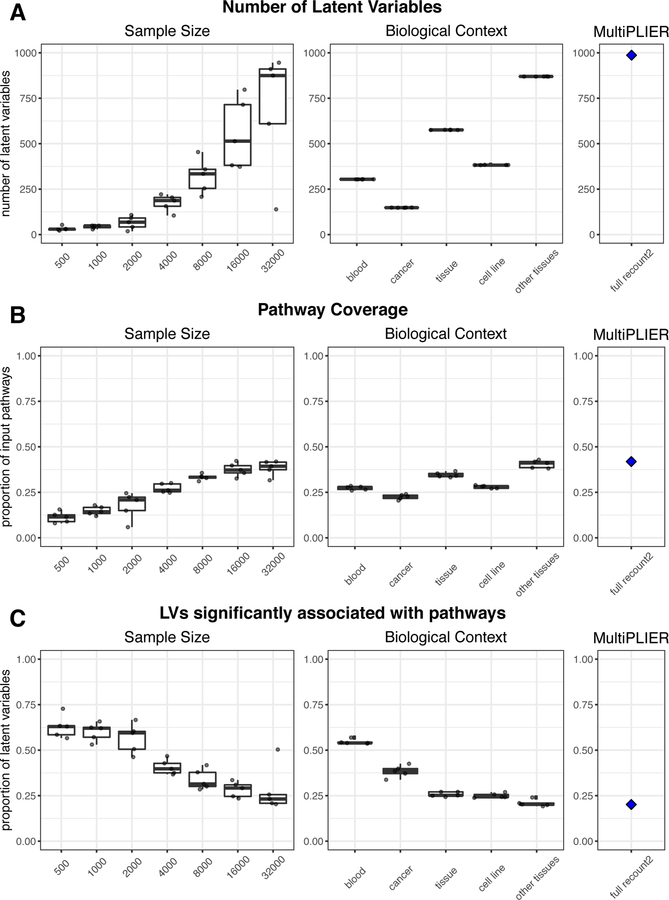
Figure 4. Subsampling of the recount2 compendium demonstrates the contribution of both sample size and breadth of biological conditions to PLIER model characteristics.
PLIER models were trained on samples randomly selected from the recount2 compendium (sample size evaluations) or on a subset of the recount2 compendium mapped to the same ontology term in MetaSRA (Bernstein et al., 2017) (biological context evaluations; see STAR Methods for the specific terms used). The training set for each repeat in the biological context evaluations is comprised of the same samples, but initialized with different random seeds. The boxplot and points in black in A-C represent 5 repeats performed for each sample size or biological context. The blue diamonds and panels labeled MultiPLIER are the values from the full recount2 PLIER model (~37,000 samples). The sample size for each biological context training set is below the biological context heatmap in panel D; the biological contexts are ordered by increasing sample size in all panels. (A) The number of latent variables (k) in a model is generally dependent on sample size. However, the biological contexts where samples are expected to be comprised of a mix of cell types (e.g., blood and tissue) have a high number of latent variable (LV) than we would expect based on the sample size experiments. (B) The proportion of pathways supplied as input to the model that are significantly associated (FDR < 0.05) with at least one latent variable, termed pathway coverage, mirrors the number of latent variables in a model. (C) The proportion of latent variables that are significantly associated (FDR < 0.05) with at least one pathway or gene set generally decreases with sample size. The exceptions are models trained on blood, which is likely the most homogeneous of the training sets, and many gene sets supplied to the models during training are immune cell related which this training set is well-suited to capture. This suggests that increasing the sample size or breadth of the training set introduces more signal that is not biologically relevant, at least with respect to the pathways that have been supplied to the model.