Figure 5. Principle component analysis (PCA) of transformed NIH/3T3 cells.
(A) PCA plot showing relationship between three MCA/TPA transformed foci, three Capan-2 exosome (ExC)/TPA transformed foci, three control foci from TPA-only treated NIH/3T3 cells, and three control foci from untreated NIH/3T3 cells. (B) PCA plot showing relationship between same three MCA/TPA transformed foci and Capan-2 exosome (ExC)/TPA transformed foci in the absence of control samples. Principle component analysis is based on comparison of exome-seq variant data using PLINK's identity-by-state (IBS) estimates.
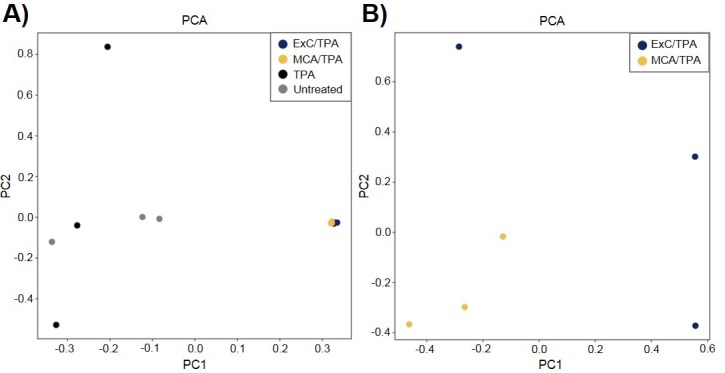
Figure 5—figure supplement 1. Variants found by Exome-sequencing analysis.