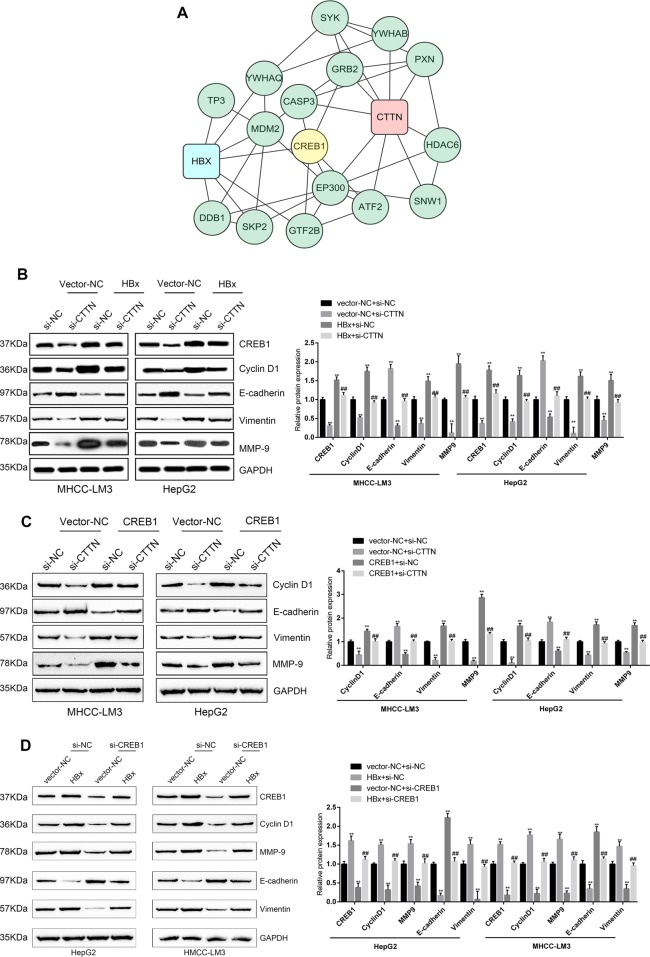
Fig. 3. The HBx-CTTN interaction modulates CREB1 and its downstream target genes.
a Analysis of HBx and CTTN protein–protein network using online databases. The network nodes represent proteins, and the connecting lines represent protein–protein interactions. b CREB1, cyclin D1, MMP-9, E-cadherin, and vimentin were detected by western blot in HepG2 and MHCC-LM3 cells. HepG2 and MHCC-LM3 cells were transfected with si-CTTN or si-NC and pcDNA3.1( + )-HBx or vector NC. si-CTTN or si-NC and pcDNA3.1( + )-HBx or vector NC were compared with the si-NC + vector-NC group. **P < 0.01. si-CTTN and pcDNA3.1( + )-HBx group were compared with HBx + si-NC group. ##P < 0.01. c The expression of EMT-associated proteins was detected in HepG2 and MHCC-LM3 cells transfected with si-CTTN or si-NC and CREB1 or vector NC. si-CTTN or si-NC and CREB1 or vector NC were compared with the si-NC + vector-NC group. **P < 0.01. si-CTTN and the CREB1 group were compared with the CREB1 + si-NC group. ##P < 0.01. d The expression of CTTN, HBx, CREB1, cyclin D1, MMP-9, E-cadherin, and vimentin was determined by western blot when HepG2 and MHCC-LM3 cells were transfected with si-CREB1 or si-NC and pcDNA3.1( + )-HBx or vector NC compared with the si-NC + vector-NC group. **P < 0.01. si-CREB1 and pcDNA3.1( + )-HBx group were compared with the HBx + si-NC group. ##P < 0.01