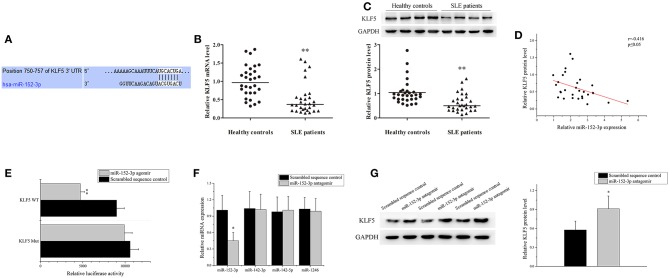
Figure 3.
Identification of miR-152-3p-targeting mRNAs in SLE B-cells. (A) The sequence of miR-152-3p-binding site in 3′-untranslated region (3′-UTR) of KLF5. (B) The KLF5 expression level in B-cells isolated from healthy controls or patients with SLE (n = 30 for each group) was analyzed by real-time PCR and normalized to GAPDH. The variables were compared using the Mann-Whitney U-test. (C) The KLF5 protein expression in B-cells isolated from healthy controls or patients with SLE (n = 30 for each group) was analyzed using western blot analysis. One representative blot is shown (upper panel). The intensity of bands was semi-quantitated and normalized to GAPDH (lower panel). The variables were compared using the Mann-Whitney U-test. (D) The correlation between miR-152-3p expression and KLF5 protein level in SLE B-cells (n = 30). (E) Relative firefly luciferase activity in Jurkat cells co-transfected with miR-152-3p agomir or scrambled sequence control together with luciferase reporter constructs containing either a WT or a mutated (Mut) KLF5 3′-UTR. Data represents the mean of three independent experiments. (F) The miR-152-3p, miR-142-3p, miR-142-5p, and miR-1246 expression levels in SLE B-cells after transfection with miR-152-3p antagomir or scrambled sequence control were analyzed by real-time PCR. All experiments were performed in triplicate. The variables were compared using the paired t-test. (G) The protein level of KLF5 in SLE B-cells after transfection with miR-152-3p antagomir or scrambled sequence control. All experiments were performed in triplicate. One representative blot is shown (left panel). The intensity of bands was semi-quantitated and normalized to GAPDH (right panel). Data represents the mean of three independent experiments. The variables were compared using the paired t-test. *P < 0.05 and **P < 0.01.