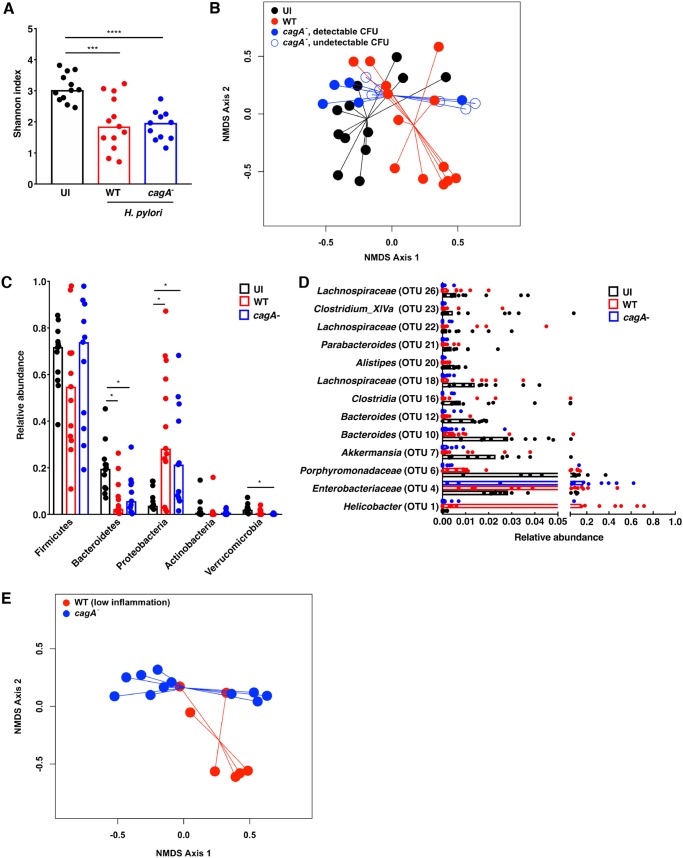
FIG 2.
H. pylori infection alters the gastric mucosal microbiota in a cagA-dependent manner. Gastric tissue from uninfected (UI) gerbils (n = 12) or gerbils infected with wild-type (WT) H. pylori strain 7.13 (n = 13) or the 7.13 cagA isogenic mutant (n = 11) was harvested 6 weeks postchallenge in linear strips, extending from the squamocolumnar junction to the proximal duodenum, and then homogenized. Microbial DNA was extracted from gastric tissue and subjected to 16S rRNA gene sequencing. (A) α-Diversity of the gastric microbiota was measured by Shannon diversity metric. ***, P < 0.0005; ****, P < 0.0001. (B) β-Diversity of the gastric microbiota was measured by Yue and Clayton’s measure of dissimilarity and is shown in a nonmetric multidimensional scaling (NMDS) plot. Blue closed circles indicate cagA isogenic mutant with detectable CFU (n = 5), while blue open circles indicate cagA isogenic mutant without detectable CFU (n = 6). Uninfected versus infected with strain 7.13, P = 0.001; uninfected versus infected with cagA mutant, P < 0.001; infected with strain 7.13 versus infected with cagA mutant, P = 0.006. (C) The relative abundances of phyla within the gastric microbiota were determined. *, P < 0.05. (D) Operational taxonomic units (OTUs) were measured by the linear discriminant analysis (LDA) effect size (LEfSe) algorithm and are shown as scatter plots. Statistical significance is indicated in the text. (E) β-Diversity of the gastric microbiota was measured by Yue and Clayton’s measure of dissimilarity and is shown in a nonmetric multidimensional scaling plot in samples stratified by the severity of gastric inflammation. Infected with strain 7.13 with low inflammation (n = 7) versus infected with cagA mutant (n = 11), P = 0.041.