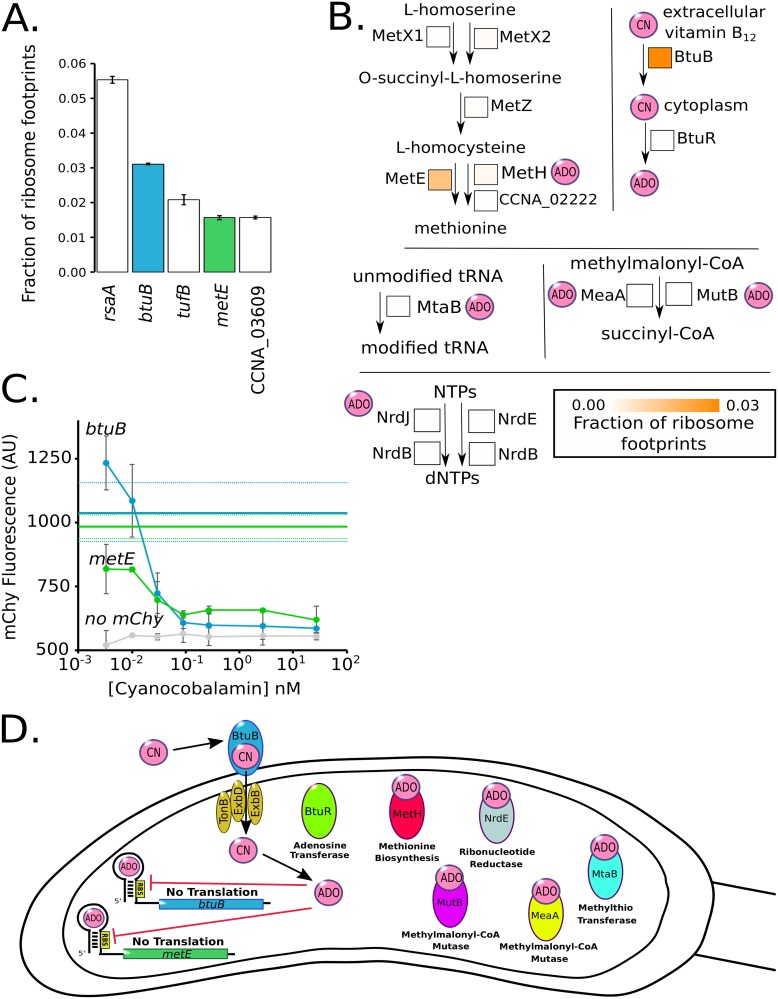
FIG 3.
C. crescentus cells are starved for B12 in laboratory growth medium. (A) Fraction of ribosome footprints for the most highly translated mRNAs in M2G. B12-related genes are colored. (B) Pathway of methionine biosynthesis, MetX1 (CCNA_03309), MetX2 (CCNA_00559), MetZ (CCNA_02321), MetE (CCNA_00515), MetH (CCNA_02221), and CCNA_02222. Pathway for B12 utilization, BtuB (CCNA_01826) and BtuR (CCNA_02321). Pathway for tRNA modification, MtaB (CCNA_03798). Pathway for nucleotide reduction, NrdJ (CCNA_01966), NrdE (CCNA_03607), and NrdB (CCNA_00261). Pathway for succinyl-CoA biosynthesis, MeaA (CCNA_03177) and MutB (CCNA_02459). Square boxes next to each enzyme contain an orange heat map which represents the fraction of ribosome footprints. ADO (adenosyl) and CN (cyano) refer to the B12 upper ligand. (C) Negative regulation by B12 riboswitches on the btuB and metE genes. Translation reporters for the btuB and metE genes fused to mCherry assayed in M2G with the indicated concentrations of B12. Error bars represent standard deviation for mCherry fluorescence in three biological replicates of the B12 dilution series (n = 3). Solid blue and green horizontal lines indicate the mChy fluorescence without vitamin B12 for btuB and metE, respectively, and dashed lines indicate the standard deviation. (D) Cartoon of B12-regulated pathways in C. crescentus. btuB and metE genes contain negative regulatory B12 riboswitches. BtuB and BtuR are part of the B12 import and utilization pathway. MetH, NrdE, MeaA, MutB, and MtaB are B12-dependent enzymes for methionine biosynthesis.