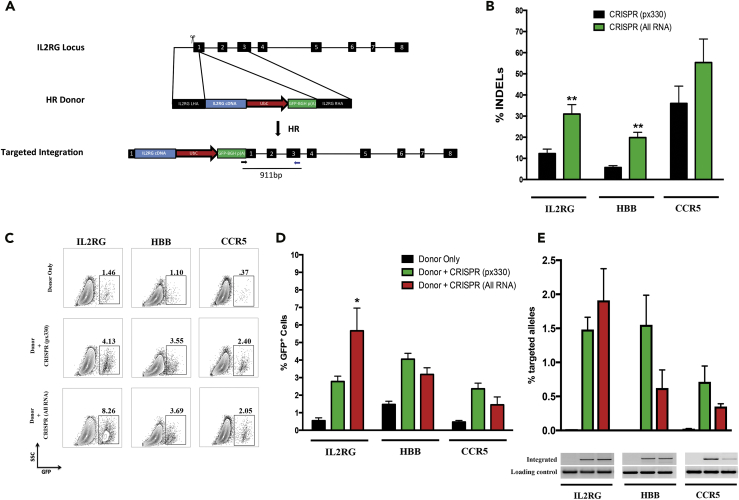
Figure 1.
CRISPR/Cas9-Mediated Homologous Recombination (HR) in NSCs
(A) The IL2RG locus was targeted for homologous recombination (HR) by creating double-strand breaks (DSBs) using Cas9 (scissors) and supplying a homologous donor template (with flanking 800-bp arms around the transgene to be inserted). Following HR, IL2RG cDNA is knocked in to the endogenous start codon with a UbC promoter driving GFP downstream to assess efficiencies of HR in human NSCs. IL2RG exons are shown in black boxes.
(B) NSCs were electroporated with 1-μg plasmid (px330) encoding Cas9 and sgRNA specific for IL2RG, HBB, or CCR5. For “All RNA” experiments, Cas9 was delivered as mRNA (5 μg), and the locus-specific sgRNAs (10 μg) were delivered with terminal chemical modifications comprising 2′-O-methyl 3′phosphorothioate (MS) or 2′-O-methyl 3′thioPACE (MSP). Following 7 days in culture, gDNA was harvested, PCR was carried out to amplify the region of interest, and INDELs were calculated using the Tracking of Indels by Decomposition (TIDE) software. (N = 2–14), **p < 0.01, Student's t test.
(C) NSCs were nucleofected as described above with either 2 μg HR donor or 2 μg HR and locus-specific CRISPR components. Locus-specific HR donor plasmids have at least 400 bp of homology flanking UbC-GFP to assess HR efficiencies. Cells were grown for at least 30 days until episomal HR donor was lost by dilution during proliferation to confirm stable expression of integrated cassettes into the IL2RG, HBB, and CCR5 loci. Representative fluorescence-associated cell sorting plots are shown, which highlight stable integration of HR donors with site-specific nucleases.
(D) HR frequencies obtained from all experiments targeting IL2RG, HBB, and CCR5 carried out as above. (N = 3–7), *p < 0.05, Student's t test.
(E) gDNA was harvested from experimental groups at day 30 post-culture to evaluate site-specific integration of HR donors on the DNA level. Digital droplet PCR (ddPCR) was used to quantify allelic integration of HR donors, (N = 3–4). Agarose gel electrophoresis of in-out PCR products confirms on-target integration of HR donors in the presence of a site-specific nuclease. Data are represented as mean ± SEM.