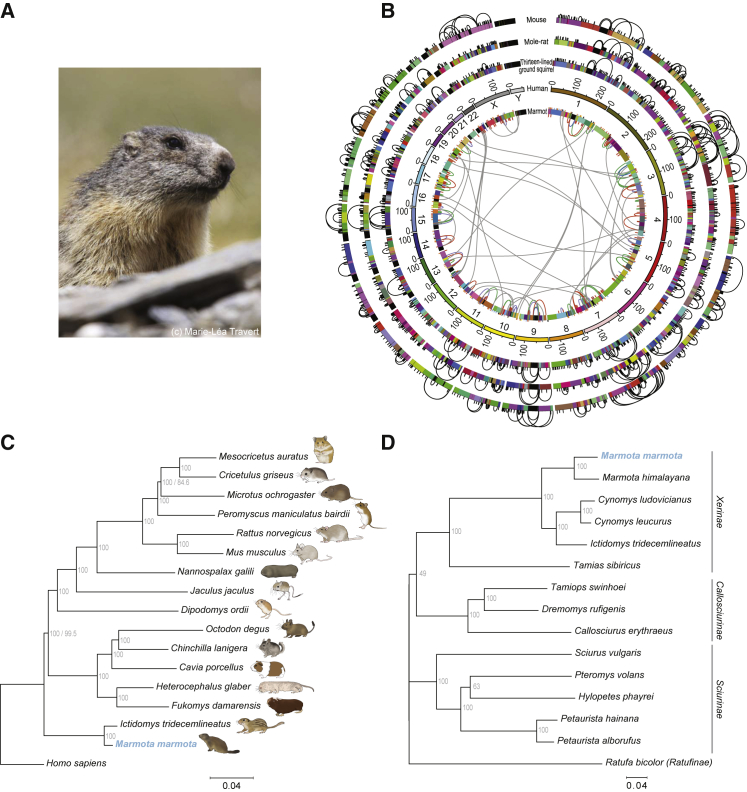
Figure 1.
A Slow Rate of Genomic Evolution and the Phylogenetic Relationship of the Alpine Marmot as Revealed by Its Nuclear and Mitochondrial Reference Genome
(A) Marmota marmota is a large, ground-dwelling, highly social rodent that has colonized high-altitude meadows across the Alps since the end of the last glaciation in the Quaternary.
(B) Collinearity of the genome assembly generated for M. marmota marmota with its close relatives, and that of other rodents. The M. marmota genome aligns to a higher fraction of the human genome (outgroup) than to its fellow rodents (i.e., mouse and mole), one of several indicators of a slower rate of genomic evolution. Here, collinear blocks in the human chromosomes are colored at random; small blocks with many colors depict lower N50 scaffold length of genome assemblies. Connections indicate collinearity breaks and block rearrangements compared to the human genome (intra-chromosomal only for the plotted rodents, except for M. marmota where interchromosomal rearrangements are plotted inside the graph). Colors indicate connections observed in M. marmota that are conserved across the sampled rodents (green; n = 72), all rodents except M. musculus (blue, n = 13), between I. tridecemlineatus and M. marmota (purple; n = 57), or that are specific to M. marmota (orange; n = 148).
(C) Reconstruction of the phylogenetic tree of Rodentia. The tree is derived from multiple whole-genome alignments of protein-coding and non-coding sequences from available rodent genomes (about 94 Mbp alignment per species). Humans are included as an outgroup. The short branch length of the Alpine marmot since the split from the last common ancestor (LCA) of primates and rodents agrees with the higher fraction of alignable genomic sequence between the Alpine marmot and human compared to Alpine marmot and mouse or mole-rat (Data S1). The scale denotes nucleotide substitutions per site.
(D) A phylogenetic tree for family Sciuridae based on their mitochondrial genomes. The scale denotes nucleotide substitutions per site.
See also Data S1 and Figures S1 and S2.