Figure 4.
The Low Genomic Diversity in the Alpine Marmot Is Explained by Its Life History and a Lack of Recovery from a Bottleneck that Coincides with the Climate Shifts of the Pleistocene
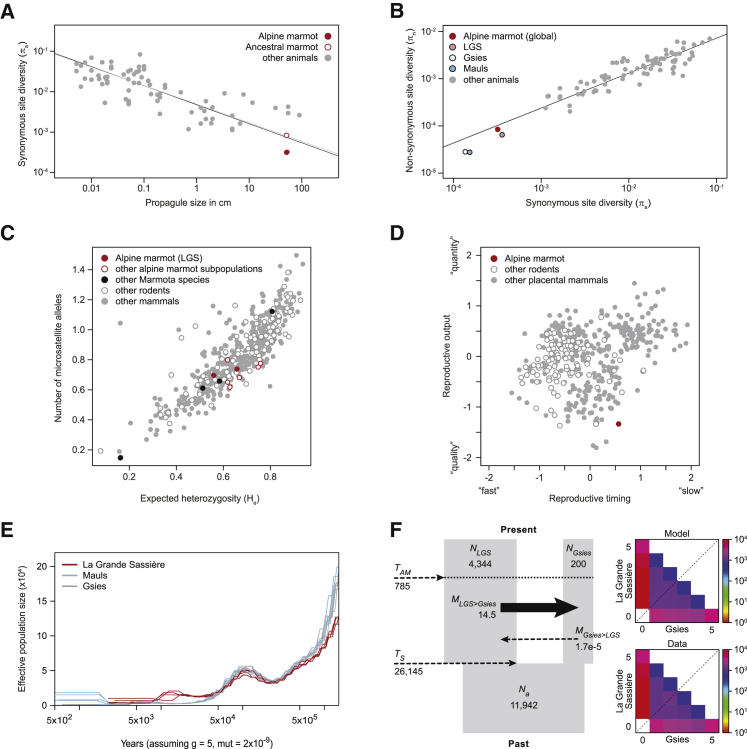
(A) The genetic diversity of the Alpine marmot is predictable from its life history. Species-wide synonymous site diversity from a wide range of animal species [24] is plotted against their “propagule size” (i.e., the size in centimeters of the dispersing life stage). The delayed dispersal of the Alpine marmot, and its large adult body size, yields a very large propagule size [25], consistent with the observed low diversity (filled red point). The diversity inferred for the ancestral marmot population at the end of the Pleistocene (empty red point; F) fits this prediction even more closely. The correlations observed are very similar whether the marmot data are excluded (gray lines) or included (black lines).
(B) The strength of purifying selection on amino acid variation in Alpine marmots corresponds to their low effective population size revealed from a pattern consistent across diverse animal species. Data from the Alpine marmot (colored points) have been added to the data of [24]. The correlations are very similar whether the marmot data are excluded (gray lines) or included (black lines).
(C) Microsatellite diversity in different Alpine marmot populations compared to many other species of mammals, including other marmot and rodent species. The number of microsatellite alleles (y axis) is plotted against the expected heterozygosity (x axis). Populations of the Alpine marmot from LGS are shown as red points, and estimates from other subpopulations of the same species, also from the French alps, are shown in as empty red bordered circles. Other species in the genus Marmota are shown as black filled circles, including the threatened M. vancouverensis, which appears at the bottom left of the graph. In difference to the genome-wide diversity, for which the Marmot is an extreme (A and B), it has a typical diversity in these sites that are characterized by a higher spontaneous mutation rate.
(D) Life history of the Alpine marmot (red point) in comparison to other Eutherian mammals (data from [26]). After correcting for body mass, much of the variance in mammalian life histories can be captured by two factors [27]: “reproductive output” (in which species vary according to their investment in offspring “quality” versus “quantity”) and “reproductive timing” (in which species vary on a “fast-slow” continuum). The Alpine marmot appears as an extreme outlier.
(E) Pairwise sequential Markovian coalescent (PSMC) analysis reveals details of the genetic past of the Alpine marmot. Evident is a decline in the LGS population after the last glacial maximum, consistent with independent evidence from the fossil record. Earlier events might suggest a longer-term decline but are more likely attributable to partial isolation between breeding populations (in which case earlier rates of coalescence reflect rates of gene flow and not Nes).
(F) The ancient migration (AM) is the most likely demographic scenario for the Gsies and La Grande Sassière (LGS) populations inferred from the joint site frequency spectrum (SFS). This model predicts that a large ancestral population split up ∼26,145 ya into two smaller daughter populations. Gene flow between these two populations ceased ∼785 ya and was strongly asymmetrical with most migrants going from the large LGS population to the very small Gsies population. The joint SFS for the LGS and Gsies populations was obtained from 178,098 SNPs (bottom right) and shows strong visual agreement with the maximum-likelihood SFS under the model (top right).
See also Table S3.