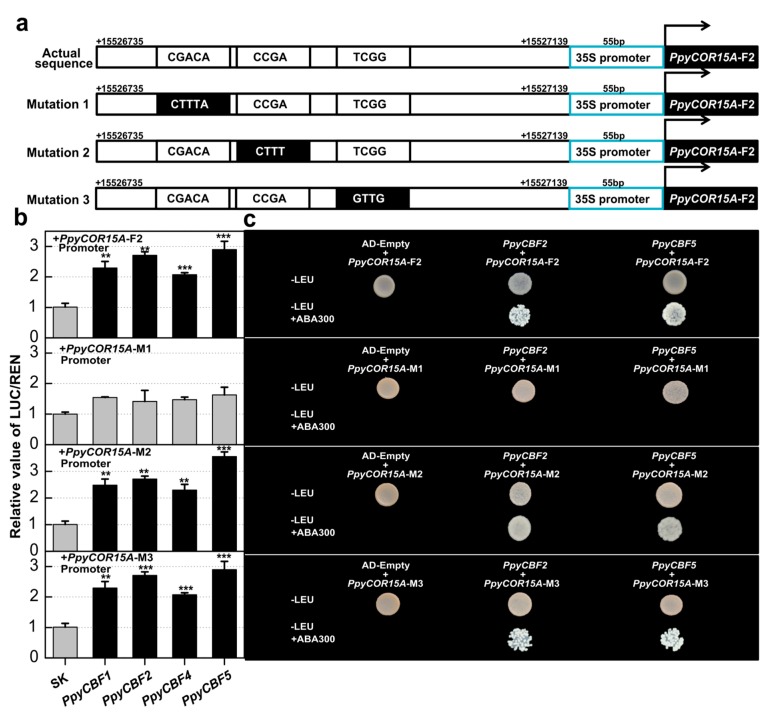
Figure 6.
PpyCBFs can also bind at TCGAC binding site in the PpyCOR15A promoter. (a) Schematic diagrams of mutations at three different motif sites for PpyCOR15A promoters, indicated with mutation 1, 2, and 3. Possible CBF-binding sites in PpyCOR15A promoter are represented with white rectangles while mutations at these sites are represented by black rectangles. (b) Dual-luciferase assays were performed with actual and mutated promoters of the PpyCOR15A promoter. The ratio of LUC/REN of the empty vector (pGreenII 0029 62-SK) plus promoter was used as the calibrator (set as 1). Three independent experiments (with minimum four replicates) were performed to verify the results. Error bars show SEs with at least four biological replicates while asterisks show significant differences with empty SK (** p < 0.01, *** p < 0.001). (c) Y1H assay was performed to check physical interaction of PpyCBFs 2 and 5 with actual and mutated promoters of PpyCOR15A. Yeast grows on synthetic dropout without leucine but having Aureobasidin A 300 (SD/−leu + ABA300) indicating the possible direct interactions.