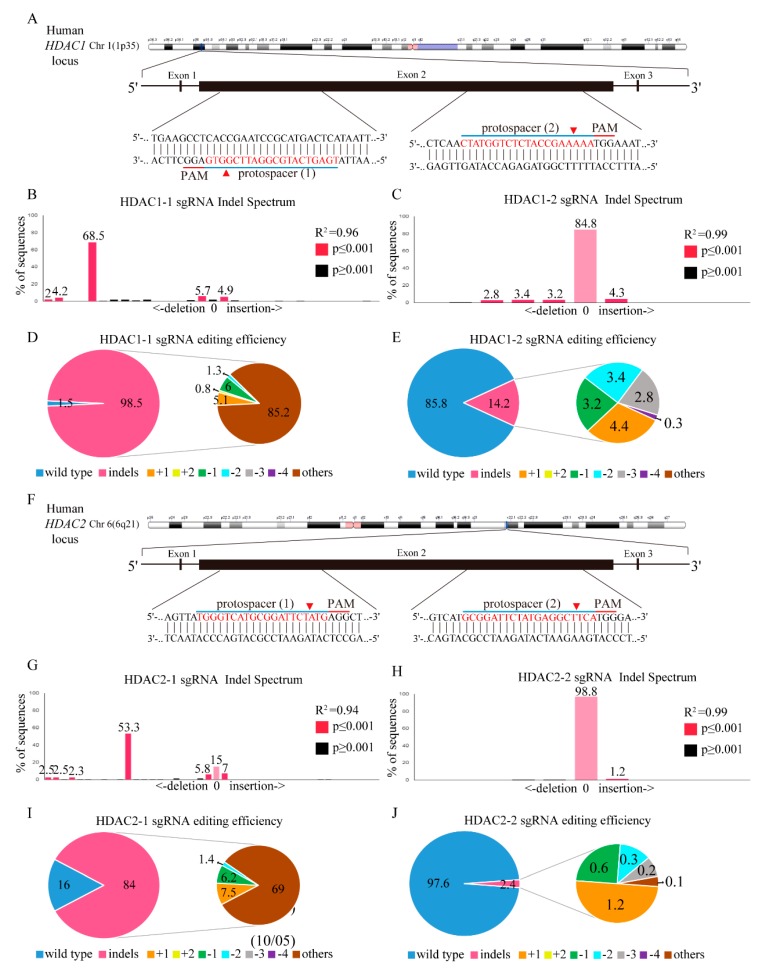
Figure 5.
HDAC1 and HDAC2 gene editing in K562 cells using the CRISPR/Cas9 system. (A) Schematic representation of the human HDAC1 DNA locus and two protospacer sequences (blue underline) for editing. The arrowhead indicates the expected Cas9 cleavage site. The protospacer adjacent motif (PAM, red underline) is the motif required for Cas9 nuclease activity. Scrambled (SC) and HDAC1 gene-edited cells were delivered to K562 cells by lentivirus. After transduction, DNA from virus-infected cells was purified and subjected to Sanger sequencing of HDAC1 exon 2. The TIDE algorithm analysis is shown for (B) HDAC1-1 and (C) HDAC1-2 gene-edited virus transfected into K562 cells compared to SC K562 cells. The pie charts show the percentages of indels in the HDAC1 gene edited by (D) HDAC1-1 and (E) HDAC1-2 gene-edited lentivirus. The gene editing efficiency of the two-gene edited virus introduced cells is presented in pink, while the two most common other mutations and +1 are presented in brown and yellow colors, respectively. (F) Schematic representation of the human HDAC2 DNA locus and two protospacer sequences (blue underline) for editing, and PAM sequences for Cas9 recognition (red underline). The arrowhead indicates the expected Cas9 cleavage site. PAM is the motif required for Cas9 nuclease activity. SC- and HDAC2-edited cells were delivered to K562 cells by lentivirus. After transduction, DNA from virus-infected cells was purified and subjected to Sanger sequencing of HDAC2 exon 2. The TIDE algorithm analysis is shown for (G) HDAC2-1 and (H) HDAC2-2 gene-edited cells virus transfected into K562 cells compared to SC K562 cells. The pie charts show the percentages of indels in the HDAC2 gene edited by (I) HDAC2-1 and (J) HDAC2-2 gene-edited lentivirus. The gene editing efficiency of the two gene-edited cells is presented in pink, while the two most common other mutations and +1 are presented in brown and yellow colors.