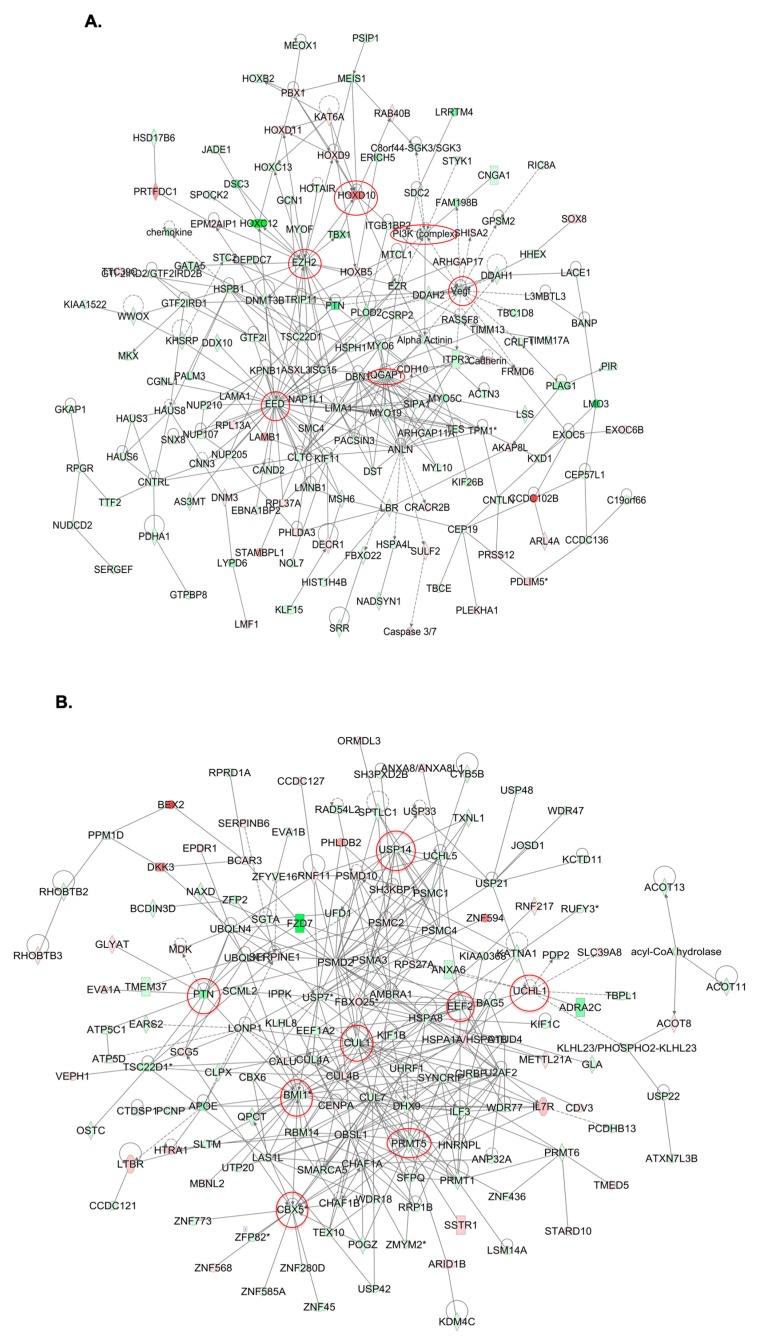
Figure 5.
Dynamic gene expression analysis in A2780s cells following both GALNT3 KO and GALNT3/T6 double KO. (A) Network analysis of dynamic gene expression in A2780s cells based on the 1.5-fold gene expression list obtained following GALNT3 KO. The five top-scoring networks of up- and downregulated genes were merged and are displayed graphically as nodes (genes/gene products) and edges (the biological relationships between the nodes). (B) Network analysis of dynamic gene expression in A2780s cells based on the 1.5-fold gene expression list obtained following GALNT3/T6 double KO. The five top-scoring networks of up- and downregulated genes were merged and are displayed graphically as nodes (genes/gene products) and edges (the biological relationships between the nodes). Intensity of node color indicates the degree of upregulation (red) or downregulation (green). Nodes are displayed using various shapes that represent the functional class of the gene product (square, cytokine, vertical oval, transmembrane receptor, rectangle, nuclear receptor, diamond, enzyme, rhomboid, transporter, hexagon, translation factor, horizontal oval, transcription factor, circle, etc.). Edges are displayed with various labels that describe the nature of the relationship between the nodes: __ binding only, → acts on. Dotted edges represent indirect interaction. Circled nodes in red represent the major gene nodes examined in this study.