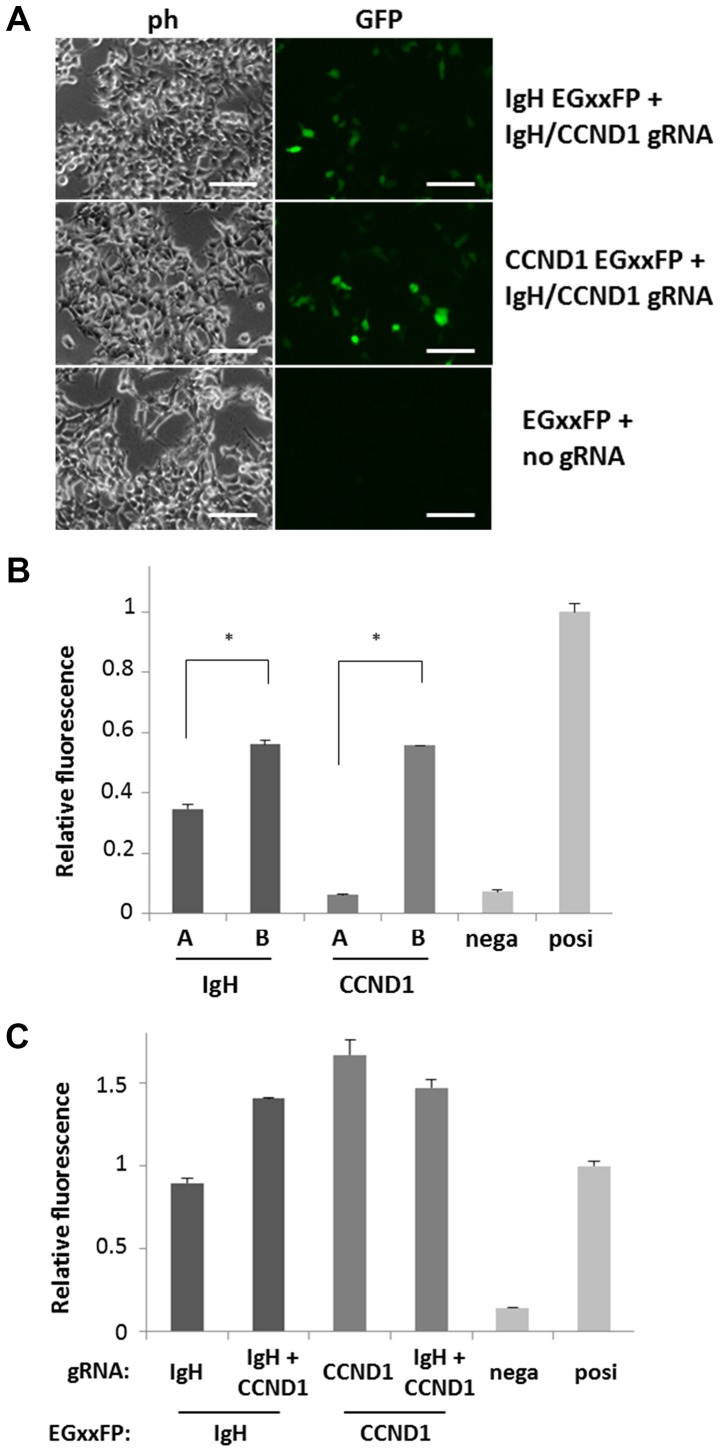
Figure 2.
Confirmation of DNA cutting activity of the gRNA candidates. (A) 293T cells were introduced with target DNA (IgH, CCND1 or empty)-containing EGxxFP and gRNA (IgH and CCND1 or empty) expressing lentiCRISPRv2 vectors. Cells with high fluorescence indicate efficient gRNA activity to cut target DNA region. Scale bars, 50 µm. (B) gRNA activity are compared among IgH and CCND1 gRNA candidates-coding lentiCRISPRv2. Bars A and B indicate the negative control (empty lentiCRISPRv2) and positive control (pX330 Cetn1), respectively. Every gRNA vector was co-transfected with the gRNA target DNA-containing EGxxFP vector, and the intensity of cell fluorescence was analyzed by flow cytometry. Relative fluorescence compared to positive control is indicated (n=3). IgH-B and CCND1-B exhibited stronger fluorescence compared to IgH-A and CCND1-A, respectively. *P<0.001, as indicated. (C) Activity of single (IgH-B or CCND1-B) gRNA and dual (IgH-B and CCND1-B) gRNA expressing vectors were compared by co-transfecting with the gRNA target DNA-containing EGxxFP vectors (indicates as EGxxFP:). Negative and positive controls are the same as those in 2B. IgH, immunoglobulin heavy chain; CCND1, cyclin D1; ph, phase contrast micrograph; GFP, green fluorescent protein; nega, negative; posi, positive.