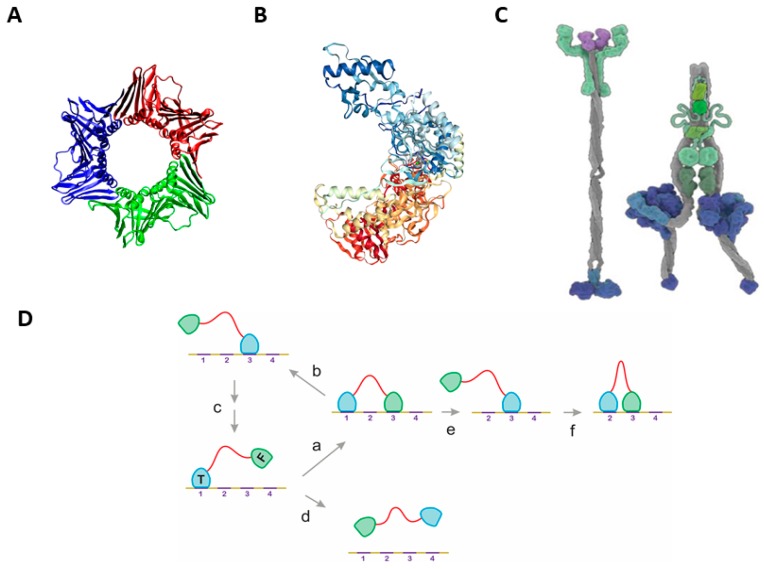
Figure 1.
Basic mechanisms of processivity. The figure illustrates the two basic types (and four subtypes) of the mechanism of processivity. The classical mechanism based on structural confinement are represented by folded proteins that either (A) completely surround their partner by an oligomeric structure of toroidal shape, such as PCNA (PDB: 1AXC) [16,17], or (B) use an asymmetric binding domain to restrict its dissociation, such as in HIV reverse transcriptase (PDB: 1REV) [18]. Basically, different mechanisms are based on spatial confinement allowed by two binding motifs connected by a long, adaptable or flexible linker, as appears in (C) the ATP-dependent dimeric mechanochemical motors kinesin-1 and dynein (adapted from [20]), or (D) monomeric processive enzymes of domain-disordered linker-domain arrangement. These types of enzymes analyzed here in detail (for cases, see Table 1) bind their polymeric substrate via two binding domains, termed “bound” or “tethered” (T) for the one that anchors the enzyme to the substrate and “unbound” or “free” (F) for the one that is in search for substrate “target” binding sites), connected by a structurally disordered linker. We show by statistical-kinetic modeling that binding via the tethering domain kinetically favors binding via the free domain (a) over full dissociation of the protein (d), which may then result in processive diffusional moves (c) or directed movements driven by energy-dependent binding and/or modification of the substrate (e,f).