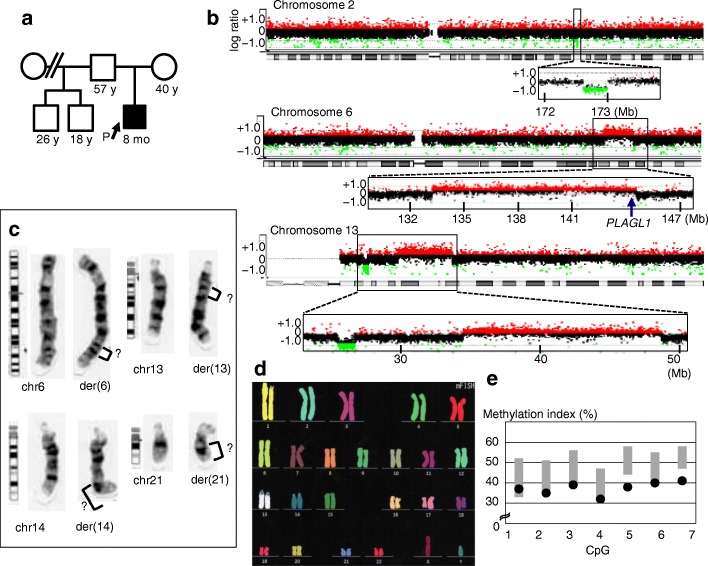
Fig. 1.
Familial information and cytogenetic findings of the patient. a Pedigree of the patient. The proband is indicated by an arrow. All other family members were clinically unremarkable. b Array-based comparative genomic hybridization showing large heterozygous copy number variations (CNVs) on chromosomes 2, 6, and 13. Large CNVs were absent from the other chromosomes. The green and red dots denote decreased (log ratio ≤ − 0.8) and increased (log ratio ≥ + 0.4) copy numbers, respectively. Numbers at the bottom of each figure indicate genomic positions based on the Human Genome Database (hg19/build 37). The position of PLAGL1, a causative gene for transient neonatal diabetes mellitus, is indicated by the blue arrow. The average log ratios for probes located in deletions and duplications were around − 1.0 and + 0.58, respectively, excluding somatic mosaicism. c G-banding of the rearranged chromosomes 6, 13, 14, and 21. No apparent abnormalities were observed on the remaining chromosomes. d Multicolor fluorescent in situ hybridization showing the lack of interchromosomal translocations. e DNA methylation analysis showing mild hypomethylation of seven CpG sites within PLAGL1. The black dots depict methylation indexes of the patient. The gray bars denote reference ranges based on data from 49 unaffected individuals