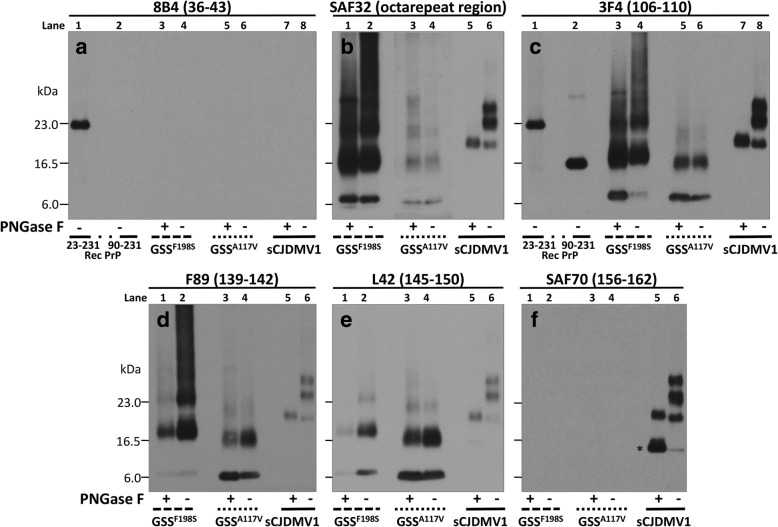
Fig. 1.
Epitope mapping combined with deglycosylation of purified resPrPD associated with GSSF198S, GSSA117V and sCJDMV1 used as control. PNGase F-treated or untreated, PK-resistant PrPD (resPrPD) purified (see Materials and Methods) from GSSF198S, GSSA117V and sCJDMV1 control were blotted and probed with the Abs to PrP denoted, with their epitopes, at the top of each panel. Panel a: Only full-length (PK-untreated) recombinant PrP (23–231) was detected by this Ab to the proximal N-terminal region confirming the absence of resPrPD with complete N-terminus. Panels b to e: Both GSSF198S and GSSA117V resPrPD conformers were selectively detected by the same Abs to PrP N- and C-terminal regions; both also showed no major variation of the resPrPD banding pattern following deglycosylation indicating that most of the resPrPD is unglycosylated in both conditions. Although overall similar, the two resPrPD profiles differed especially in the higher molecular weight region suggesting a distinct repertoire, or relative proportions, of polymers in the two GSS variants. The GSS banding patterns clearly differed from the three-band pattern of sCJDMV1, two of which are glycosylated (See Results for detailed description). Panel c includes the bands of both 23–231 and 90–231 recombinant PrP which have been used as molecular weight markers. In b the portion of the panel with the GSSA117V samples required longer exposure. In panel f, * indicates the 13 kDa component of the glycosylated and anchor bearing PrP 12/13 C-terminus fragments with N-termini at residues 162–167 and 154–156, respectively [24, 41]