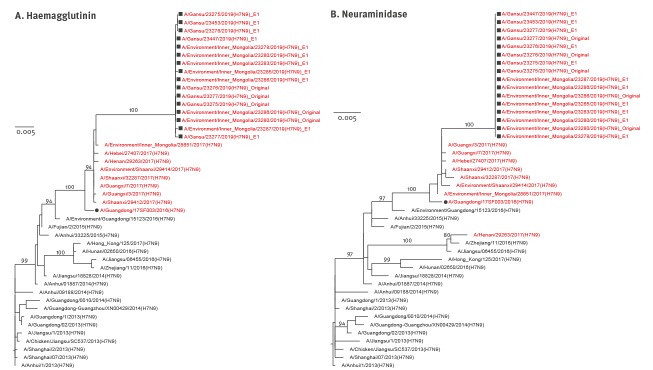
Figure 1.
Phylogenetic analyses of the (A) haemagglutinin and (B) neuraminidase gene segments of HPAI H7N9 viruses recovered from an infected patient and from environmental samples collected nearby, Inner Mongolia Autonomous region, China, April 2019
HPAI: highly pathogenic avian influenza; MEGA: Molecular Evolutionary Genetics Analysis; MUSCLE: MUltiple Sequence Comparison by Log- Expectation.
Multiple sequence alignments were performed with the MUSCLE programme using MEGA software version 7. Maximum likelihood trees on (A) haemagglutinin and (B) neuraminidase genes were conducted using the general time reversible + Γ nt substitution model with 1,000 bootstraps, respectively. HPAI H7N9 viruses are highlighted in red. The investigated viruses and the candidate HPAI H7N9 vaccine strain are shown with a solid square and circle, respectively. Bootstrap values higher than 60 are shown. Horizontal distances are proportional to genetic distance, as shown by the scale bars.