Fig. 2.
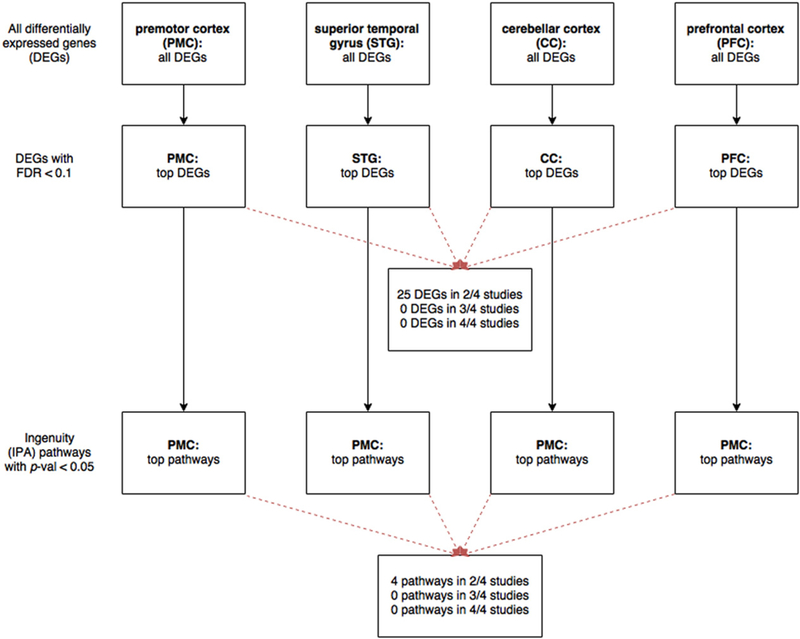
Workflow of analysis to identify differentially expressed genes with FDR< 0.10, and their associated pathways.We identified four studies by our systematic review: Chen et al. (inpremotor cortex, PMC), Pietersen et al. (superior temporal gyrus, STG),Mudge et al. (cerebellar cortex, CC), and Iwamoto et al. (prefrontal cortex, PFC). After extracting the expression data, we analyzed differentially expressed genes (DEGs) mediated by clozapine vs. other atypical antipsychotic medications. For each study, we ranked the DEGs by false discovery rate (FDR) and selected those with FDR < 0.10.We also performed Ingenuity pathway analysis (IPA) on the geneswith FDR b 0.10 in each study, and identified canonical pathwayswith p-value <0.05
