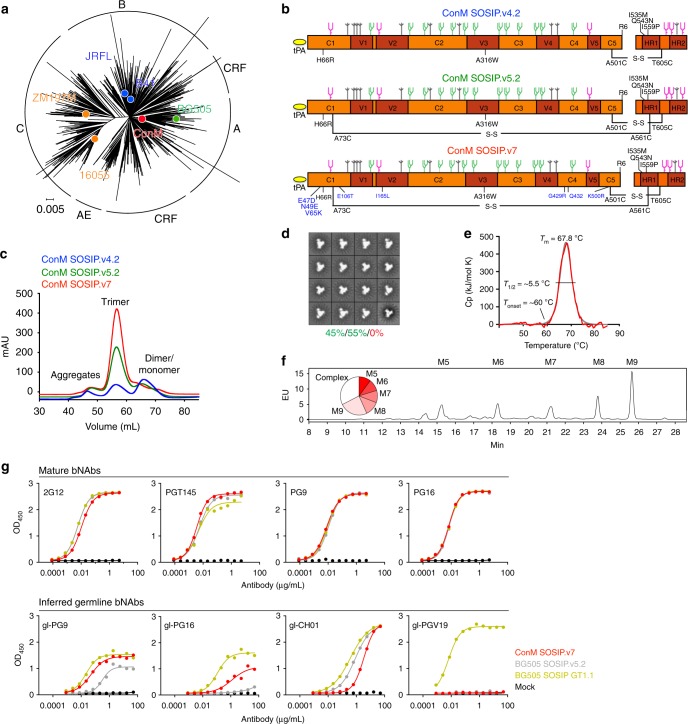
Fig. 1.
Design, screening, and biophysical characterization of ConM SOSIP trimers. a Maximum likelihood tree generated with all Env sequences of the M group from the Los Alamos HIV database webalignment 2011 (n = 3654) and the parental Env sequences of five previously described SOSIP trimers4, 17, 72, 81 and ConM. Different subtypes and genetic distance bar (substitutions per sequence position) are indicated. b Linear representation of the three ConM SOSIP variants. Top: design of SOSIP.v4.2 with the SOS-bond (A501C-T605C), improved furin cleavage site (R6), IP (I559P), trimer-stabilizing mutations (H66R and A316W), and improved trimerization mutations (I535M and Q543N)4, 16. Middle: design of SOSIP.v5.2 with an extra intermolecular disulfide bond (A73C-A561C)5. Bottom: design of SOSIP.v7 with TD8 mutations in blue17. The assignment of glycans was based on the analysis of BG505 SOSIP.664 reported in ref. 41 but may be different for ConM SOSIP trimers. c Size exclusion chromatographic profiles of 2G12-purified ConM SOSIP trimers (untagged) expressed in 293S cells on a Superdex200 16/60 column. d 2D class average of negative-stain electron microscopic analyses of PGT145-purified ConM SOSIP.v7 trimer (D7324-tagged) expressed in 293F cells. Percentages of closed and open native-like are depicted in green and non-native in red. e Thermostability derived from differential scanning calorimetry (DSC) of PGT145-purified ConM SOSIP.v7 trimer (D7324-tagged) expressed in 293F cells. The DSC parameters midpoint of thermal transition (Tm), starting temperature of unfolding (Tonset), and width of the transition at half peak height (T1/2) are indicated. f Hydrophilic interaction liquid chromatography-ultra performance liquid chromatographic spectra of N-linked glycans derived from PGT145-purified ConM SOSIP.v7 trimer (D7324-tagged) in 293F cells. Peaks corresponding to oligomannose glycans (Man5–9GlcNac2) are labeled M5–M9. The smaller peaks that are not highlighted correspond to complex or hybrid-type glycans. The relative abundances of M5–M9 and complex glycans are summarized in a circle diagram. g Ni-NTA-capture enzyme-linked immunosorbent assay with PGT145-purified ConM SOSIP.v7 trimer and the comparator trimers BG505 SOSIP.v5.2 and GT1.1 (all His-tagged) expressed in 293F cells against a panel of (gl-)bNAbs