Abstract
Taste receptors including calcium sensing receptor (CaSR) are expressed in various animal tissues, and CaSR plays important roles in nutrient sensing and the physiology, growth, and development of animals. However, molecular distribution of porcine CaSR (pCaSR) in different tissues, especially along the longitudinal axis of the digestive tract in weaned piglets, is still unknown. In the present study, we investigated the distribution and localization of pCaSR in the different tissues including intestinal segments of weaned piglets. Six male pigs were anesthetized and euthanized. Different tissues such as intestinal segments were collected. The pCaSR mRNA abundance, protein abundance, and localization were measured by real-time PCR, Western blotting, and immunohistochemistry, respectively. The mRNA and protein of pCaSR were detected in the kidney, lung, liver, stomach, duodenum, jejunum, ileum, and colon. The pCaSR mRNA was much higher (five to 180 times) in the kidney when compared with other tissues (P < 0.05). The ileum had higher pCaSR mRNA and protein abundances than the stomach, duodenum, jejunum, and colon (P < 0.05). Immunohistochemical staining results indicated that the pCaSR protein was mostly located in the epithelia of the stomach, duodenum, jejunum, ileum, and colon. These results demonstrate that pCaSR is widely expressed in different tissues including intestinal segments in weaned piglets and the ileum has a higher expression level of pCaSR. Further research is needed to confirm the expression of CaSR in the different types of epithelial cells isolated from weaned piglets and characterize the functions of pCaSR, its potential ligands and cell signaling pathways related to CaSR activation in enteroendocrine cells and potentially in enterocytes.
Keywords: calcium sensor receptor, gene expression, intestine, nutrient sensing, weaned piglets
INTRODUCTION
The gut operates not only as an organ to digest and absorb nutrients but it also acts as a chemosensory system mediating the orchestration of physiological and metabolic responses (Reimann et al., 2012; Burrin et al., 2013; Ipharraguerre et al., 2013; Kaji et al., 2013). The gut chemosensing system is considered to be characterized by a network of G protein-coupled receptors (GPCRs) that is associated with the regulation of nutrient absorption and gut function (Liu et al., 2013). Identification of specific GPCR in the gut and their ligands can provide novel targets not only for improving gut growth and development but also for the treatment of diabetes, acid reflux, mucosal injury, inflammatory bowel disease, and obesity (Symonds et al., 2015; Cheng, 2016; Owen et al., 2016). Antibiotics are widely used to maintain health and productivity within the animal production chain (Hassan et al., 2018). However, the consumption of antibiotics in food animal production may lead to the spread of antibiotic resistant pathogens, posing a significant public health threat (Yang et al., 2015; Van Boeckel et al., 2015). Therefore, new technologies are needed.
Modulating gut chemosensing has become an area of future interest to further develop novel therapeutic strategies against weaning-induced enteric dysfunction in pigs (Liu et al., 2013). As a part of gut chemosensing, taste receptors are GPCRs that play crucial roles in nutrient sensing in the intestine. The receptors for sweet, umami, kokumi (pCaSR), and fatty acid have been identified and characterized in the pig (Tedo et al., 2011; Maruyama et al., 2012; Mace and Marshall, 2013; Yang et al., 2013; Song et al., 2015). However, molecular distribution of pCaSR in different tissues, especially along the longitudinal axis of the digestive tract in weaned piglets, is still unknown. We hypothesized that pCaSR has a similar molecular structure with other mammalian CaSR and is widely expressed in different tissues including intestinal segments of weaned piglets. Thus, the objective of this study was to investigate the distribution and localization of pCaSR in different tissues, especially along the longitudinal axis of the digestive tract in weaned piglets.
MATERIALS AND METHODS
Database Information Collection
The sequences of pCaSR gene and protein investigated in this study were collected from GenBank (https://www.ncbi.nlm.nih.gov/gene/100520980), and then were analyzed by using Ensembl (http://uswest.ensembl.org/Sus_scrofa/Gene/Summary?g=ENSSSCG00000011878;r= 13:138280364-138364953), and the pCaSR Uni ProtKB/SWISS-Prot database information were obtained from (http://www.uniprot.org/uniprot/O62714). The exon/intron organization of the pCaSR gene was drawn by using Illustrator for Biological Sequences (Liu et al., 2015). Secondary structure model of the pCaSR was constructed by Protter (http://wlab.ethz.ch/protter/start/). The hydropathy plot analysis of the pCaSR protein was conducted by using ExPASy-Protscale (https://web.expasy.org/protscale/). The predicted 3D structure model of the pCaSR AAs sequences was built by Swiss-model (https://swissmodel.expasy.org/interactive). The homologies of the CaSR mRNA and AA sequences among pig and other five species such as Homo sapiens, Mus musculus, Gallus gallus, Rattus norvegious, and Copra hircus were analyzed by Clustal W from BioEdit version 7 program (http://www.mbio.ncsu.edu/bioedit/bioedit.html). The phylogenetic tree constructed by the maximum-likelihood model constructed with 1,000 bootstrap replicates using MEGA 6.0 software (Tamura et al., 2013).
Animals and Sample Collection
The experimental protocol was reviewed and approved by the Animal Care Committee of the University of Manitoba and the pigs were cared for in accordance with the Canadian Council on Animal Care guidelines (CCAC, 2009). Six male pigs ([Yorkshire-Landrace] × Duroc, weaned at 21 ± 2 d) at the age of 28 d were anesthetized by an intramuscular injection of ketamine: xylazine (20:2 mg/kg BW) and euthanized by i.v. injection of sodium pentobarbital (50 mg/kg BW) (Aluko et al., 2017). Different tissue samples such as kidney, lung, liver, tongue, stomach, duodenum, jejunum, ileum, and colon were collected. Each tube, which contained ~15 g of tissue, was tightly capped, and immediately frozen in liquid nitrogen (N2). The frozen samples were subsequently pulverized to be homogenous with a mortar and a pestle (Fisher Scientific, Burlington, USA) under liquid N2, and stored at −80 °C until required for analysis (Yang et al., 2016b). A 5-cm section of each tissue was fixed by immersion in 10 % neutral buffered formalin (Fisher Scientific) for histology and immunohistochemistry analyses (Yang et al., 2011).
Total RNA Isolation, cDNA Synthesis, and Real-Time PCR Analyses
Total RNA was isolated from pulverized tissue samples using a RNAqueous Total RNA Isolation Kit (Invitrogen, Carlsbad, Canada) according to manufacturer’s instructions. The total RNA concentration was determined by measuring the absorbance at 260 nm using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Ottawa, Canada) and RNA integrity was verified by visualization in an agarose gel. One microgram of total RNA was reverse transcribed into cDNA using an iScript cDNA Synthesis Kit (Bio-Rad Laboratories Ltd., Mississauga, Canada) following the manufacturer’s instruction. Real-time PCR was performed using a Thermocycler (Bio-Rad Laboratories Ltd.) and SYBER Green Supermix (Bio-Rad Laboratories Ltd.). The specific primers of pCaSR were designed based on the published cDNA sequence in the Genbank. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control to normalize the input amount of RNA. The forward (F) and reverse (R) primers for pCaSR were: CaSR-F (5′-GCCAAAGATCAGAACCTAG-3′) and CaSR-R (5′-GCTGTTTATTTCCTCTATG-3′), and primers for GAPDH were GAPDH-F: (5′-GTGAACGGATTTGGCCGC-3′) and GAP DH-R (5′-AAGGGGTCATTGATGGCGAC-3′). The following protocol was used: 95 °C for 5 min followed by 40 cycles of 95 °C for 15 s and 58 °C for 15 s and 72 °C for 30 s. Melting curve analysis was conducted to confirm the specificity of each product, and the size of products were verified on 1% agarose gels in Tris acetate–EDTA buffer. The pCaSR PCR product was purified using a Thermo Scientific GeneJET Gel Extraction Kit (Thermo Fisher Scientific) according to the manufacturer’s instruction, and the identity of the pCaSR product was confirmed by sequencing at the Research Institute in Oncology and Hematology at the University of Manitoba. Data were analyzed using the 2−ΔΔCT method (Livak and Schmittgen, 2001) to calculate the relative fold change of target gene.
Western Blotting Analyses
Total protein was extracted from the stomach, duodenum, jejunum, ileum, and colon using a total protein extraction kit according to the manufacturer’s instruction (Thermo Fisher Scientific). Protein concentration was determined by using a commercial BCA protein assay kit (Thermo Scientific) with bovine serum albumin (fraction V) as a standard. The protein levels of pCaSR and β-actin were determined by Western blotting analyses (Yang et al., 2016a). In brief, proteins (25 μg) were separated by electrophoresis in an 8% SDS polyacrylamide gel. Proteins were transferred from the gel onto a nitrocellulose membrane followed by incubation with mouse anti-CaSR primary antibodies (ab19347, Abcam, Cambridge, MA) that were diluted in 5% milk in TBST (1:1,000) for 2 h at room temperature. Membranes were subsequently probed for 1 h at room temperature with an HRP-conjugated anti-mouse IgG antibody (#7076, Cell Signaling, Danvers, MA) that was diluted in 5% milk in TBST (1:2,000). Then the membranes were visualized using the Luminata Crescendo chemiluminescent HRP detection reagent (Millipore (Canada) Ltd., Etobicoke, ON, Canada). To ensure equal protein loading, the same membranes were reprobed with a rabbit anti-β-actin monoclonal antibody (1:2,000; #4967, Cell Signaling) and then incubated with an anti-rabbit IgG antibody (#7074, Cell Signaling) for 1 h at room temperature, and visualized using the Luminata Crescendo chemiluminescent HRP detection reagent (Millipore (Canada) Ltd.). Photographs of the film were scanned, and densitometry was quantified. Western blotting analyses were all performed in duplicate for each sample.
Immunohistochemistry
Tissues were collected, fixed in 10% buffer saturated formaldehyde, and embedded in paraffin. Tissue sections (0.5 μm) were cut using a microtome (Leica, Richmond Hill, Canada), collected on Super Frost Plus slides (Thermo Fisher Scientific). Slides were placed in xylene and gradient ethanol, respectively, for deparaffinization and rehydration. Slides were treated with a boil in 10 mM sodium citrate buffer (pH at 6.0) and remained at a sub-boiling temperature for 10 min using a microwave oven at pressure, then cooled on bench top for 30 min to unmask antigen. After being treated with 3% hydrogen peroxide plus 10% methanol for 10 min to block endogenous peroxidases, slides were incubated with the avidin solution and biotin solution (Avidin/Biotin Blocking system, Biolegend, San Diego) according to the manufacturer’s instruction to block endogenous avidin/biotin. Following 60-min procedure to block unspecific binding with 20% normal goat serum (Jackson ImmunoResearch Laboratories, West Grove, PA), slides were incubated with primary antibodies (ab19347, Abcam) at a dilution of 1:50 at 4 °C overnight and then being added a Goat Anti-Mouse IgG (H+L) (Jackson ImmunoResearch Laboratories) secondary antibody at a dilution of 1:500 for 1 h at room temperature, and peroxidase-conjugated streptavidin (1:500, Jackson ImmunoResearch Laboratories) at room temperature for 30 min. All slides were counterstained with hematoxylin (Mayer’s hematoxylin, Saint Louis). For negative control, nonimmune mouse serum diluted to a protein concentration similar to that of the diluted mouse anti-CaSR antibody was substituted for the primary antibody. Slides were photographed using a Zeiss Axio Scope.A1 (Car-Zeiss Ltd, Toronto, ON, Canada).
Statistical Analysis
Data obtained from the real-time RT-PCR and the Western blotting analyses were presented as means ± SEM and subjected to the analysis of variance using SAS (the SAS Institute, Cary, NC). Comparisons of the molecular endpoints among the different tissues were further conducted by using the Tukey’s multiple comparisons of the SAS. Statistical significance was considered at P < 0.05. The figures were made using the GraphPad Prism 7 (GraphPad Software, La Jolla).
RESULTS
Structure Features of pCaSR Gene and Protein
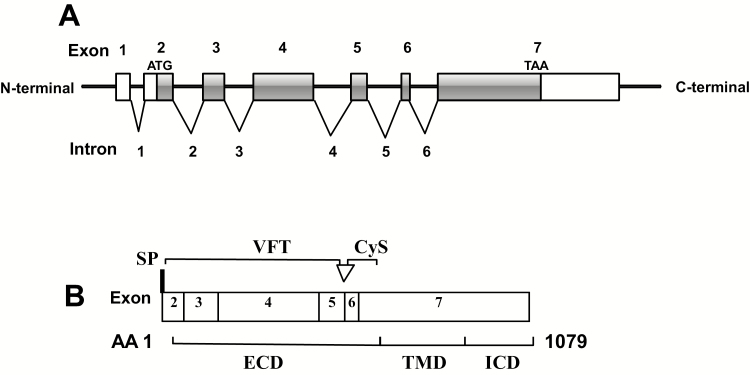
From the pCaSR gene information provided by the Ensembl genome database, it showed that pCaSR gene is located in the porcine chromosome 13q (138, 150, 871 to 138, 310, and 872) and spans ~104 kb. Also, the T-cell antigen CD86 gene lies upstream and the family with sequence similarity 162 members A gene (FAM162A) downstream of the pCaSR gene. As shown in Figure 1A, the pCaSR gene has seven exons and six introns. The full-length pCaSR coding mRNA consists of 3,240 nucleotides, and the pCaSR protein coded by exon 2 to exon 7 is comprised of 1,079 AAs. For pCaSR, the 5′ UTR upstream of the ATG translation initiation codon is encoded by exon 2 and consisted of 228 nucleotides, and the exon 7 encodes 1,508 nucleotides 3′ UTR downstream of the TAA translation stop codon. Also, pCaSR exists an alternative RNA transcript which has extra 30 bases.
Figure 1.
A schematic diagram of the pCaSR gene exon/intron and protein domains. Exons were drawn to the scale, but introns were not. (A) There are seven exons and six introns in the DNA of the pCaSR gene. Blank bars (part of exon 2 and 7) represent exons which are not involved in mRNA translation and gray bars represent mRNA coding regions including part of exon 2, exons 3 to 6 and part of exon 7). ATG, initiation codon; TAA, stop codon. (B) pCaSR protein contains 1,079 AA residues. Open arrowhead: additional 10 AA which is associated with alternative splicing. SP, signal peptide; VFT, venus flytrap domain; Cys, cysteine rich domain; ECD, extracellular domain; TMD, transmembrane domain; ICD, intracellular domain.
According to the manual assertion inferred from sequence similarity to human CaSR protein (Figure 1B), it is suggested that pCaSR also contains a 19 AA N-terminal signal peptide (1 to 19), a 593 AA extracellular domain (ECD) (20 to 612) comprising a 507 AA venus flytrap (VFT) domain (22 to 528), seven transmembrane domains (TMDs) containing 250 AAs (613 to 862) and a 217 AA intracellular domain (ICD) (863 to 1079). In addition, the ECD is linked with the ICD by a 57 AA cysteine-rich (Cys) domain (542 to 598).
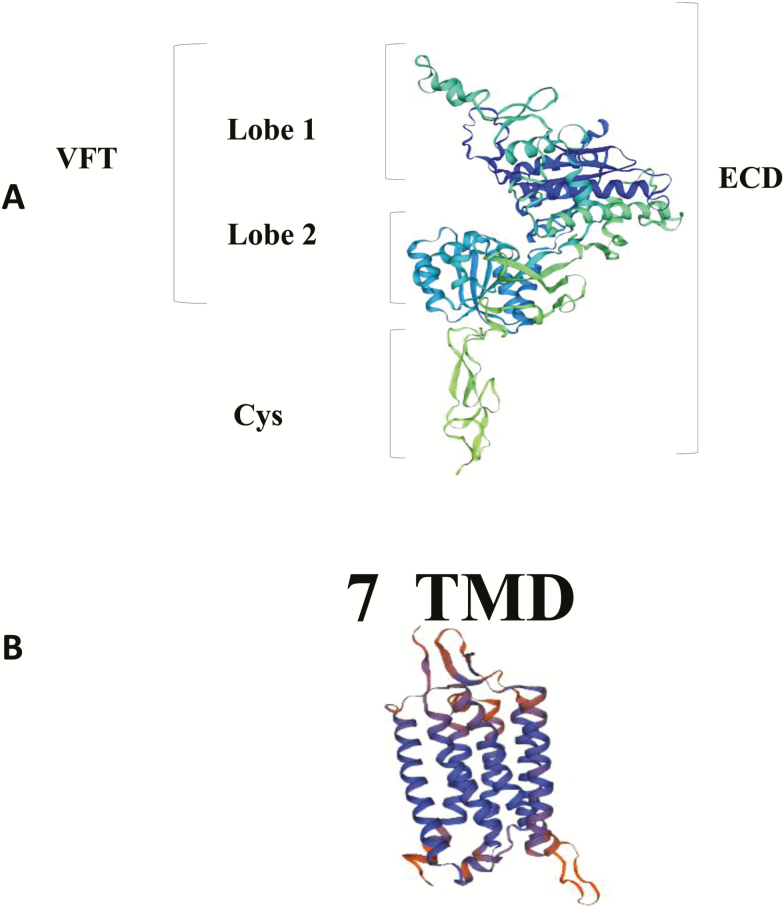
From three-dimensional protein homology model of pCaSR ECD which was constructed based on the crystal structure of human CaSR using SWISS-MODEL (Figure 2A), it showed that the ECD is composed of a VFT domain formed by two lobes (lobes 1 and 2) and a Cys domain. Meanwhile, according to the homology model of TMD of the pCaSR which was constructed based on the metabotropic glutamate receptor 1 using SWISS-MODEL (Figure 2B), it showed TMDs consist of seven α-helices.
Figure 2.
3D homology models of the pCaSR extracellular domain (ECD) (A) and of the seven transmembrane domain (TM) module helices (B). The pCaSR ECD homology model and the helices of pCaSR seven TM module were modeled on a human calcium sensing receptor template (Geng et al., 2016) and metabotropic glutamate receptor 1(Wu et al., 2014) using the SWISS-MODEL, respectively.
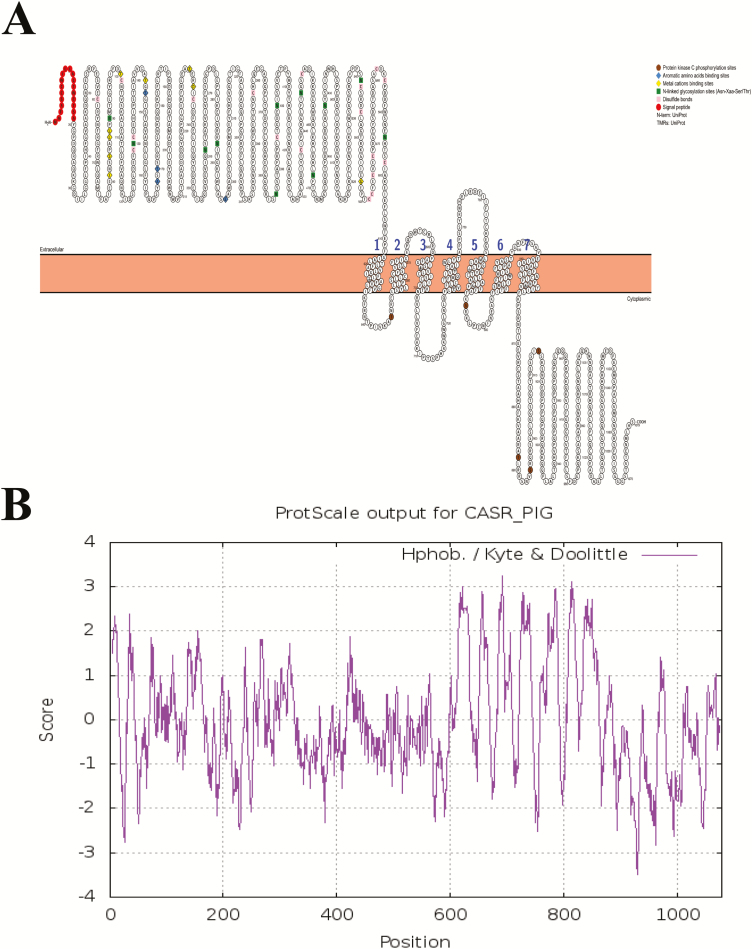
Furthermore, based on the manual assertion inferred from the combination of experimental and computational evidence of human CaSR (Geng et al., 2016; Zhang et al., 2016), putative ligand-binding sites of pCaSR were also shown in Figure 3A, containing nine calcium-binding sites located within pCaSR ECD (positions 81, 84, 87, 88, 100, 145, 231, 234, and 557), as well as four aromatic AA-binding sites (positions 147, 168, 170, and 297). There are 11 heavily glycosylated and conserved N-linked glycosylation sites (Asn-Xaa-Ser/Thr) and 18 disulfide bonds within ECD. Five conservative protein kinase C phosphorylation sites are present within TMDs and ICD. Specifically, two protein kinase C phosphorylation sites were found in the first and third TMD loops within the carboxy-terminal domains.
Figure 3.
Schematic of the pCaSR secondary structure model and ligands binding sites and transmembrane domains (A) and hydropathy plot analysis of the 1,079 AA in pCaSR protein using the scale Hphob./Kyte & Doolittle (B).
As shown in Figure 3B, it is a schematic representation of pCaSR primary structural features. The hydropathy profile and general topology of pCaSR resemble human CaSR and the other mammalian homologs. It showed that pCaSR contains a large hydrophilic amino-terminal ECD, seven hydrophobic TMDs, and a carboxy-terminal hydrophilic ICD.
CaSR mRNA and AA Sequences Alignment and Phylogenetic Analysis
The pCaSR mRNA sequence has 80.2% to 92.6% homology with the CaSR mRNA sequence of chicken, rat, mouse, human, and goat, while the pCaSR AA sequence has 87.4% to 96.6% homology with the CaSR AA sequence of these species. The pCaSR AA sequence has 96.6% homology with the Campra hircus CaSR (KF006348). Compared with the AA sequences of human CaSR (U20759), a total of 58 residue variances were identified in the pCaSR, containing 57 AA variances and a proline insertion at position 953 within ICD. In particular, three AA variances appeared within the signal peptide (Y4→S, V9→I, H16→C) and TMD (N583→D, V740→A, Q857→V), respectively, and 17 variances were observed within ECD (D50→N, K52→E, L92→M, H254→Q, Q309→E, K335→Q, R340→S, PV369-370→TT, S380→G, D382→G, F384→I, I409→M, N471→S, C483→Y, V486→A, N583→D), while there were also 35 variances which were identified within ICD (K897→Q, Q932→K, QEQ941-943→HVP, Q945→P, PLTL947-950→APST, 953→P, QRS955-957→PQL, N986→S, M988→T, Q996→K, SS1004-1005→NN, T1007→A, P1011→A, T1020→A, LD1024-1025→AE, F1036→S, QR1041-1042→HH, V1045→M, L1051→M, V1057→M, S1059→N, Q1061→R, VVN1076-1078→MLH).
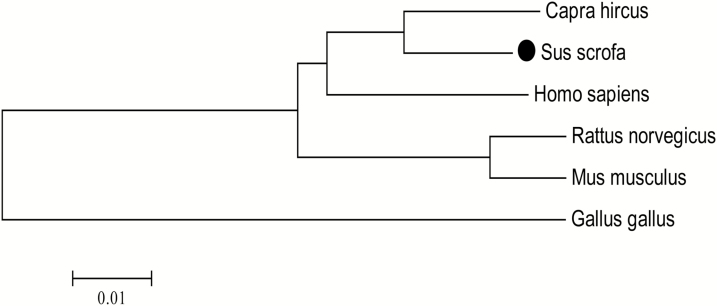
As shown in Figure 4, the phylogenetic tree of CaSR showed that these six CaSR protein sequences were clustered into two groups: avian and mammal. The pCaSR was located in vertebrates group, and brunches of pig and goat were clustered firstly, then clustered with human and rat, finally clustered with chicken. The evolutionary relationships revealed that the pCaSR is closer to the goat compared with the other four species.
Figure 4.
A phylogenetic tree of the AA sequences of chicken, pig, rat, mouse, human, and goat CaSR. The tree was constructed by the neighbor-joining method based on the Poisson correction model with 1,000 bootstrap replicates using MEGA 6.0 software. The bar (0.01) indicates the genetic distance. GenBank accession numbers: Sus scrofa (GenBank ID: NM_001278748), Capra hircus (GenBank ID: KF006348), Homo sapiens (GenBank ID: U20759), Rattus norvegicus (GenBank ID: AAC52195), Mus musculus (GenBank ID: AAD28371), and Gallus gallus (GenBank ID: XP_416491.5).
Real-Time PCR Analyses of pCaSR mRNA Abundance in Different Tissues
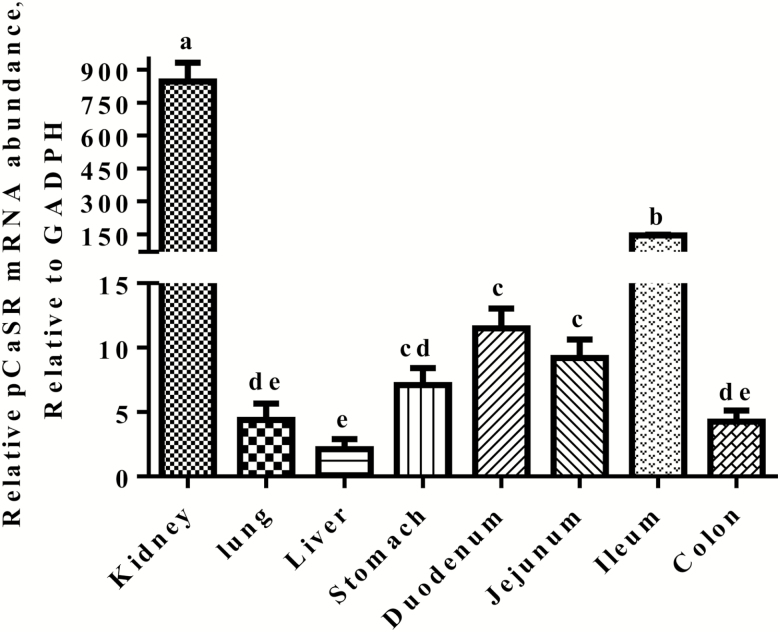
As shown in Figure 5, the pCaSR mRNA was detected in the different tissues such as kidney, lung, liver, stomach, duodenum, jejunum, ileum, and colon in weaned piglets. The pCaSR mRNA expression level was much higher (five to 180 times) in the kidney when compared with other tissues (P < 0.05). The ileum had higher (four to 18 times) pCaSR mRNA abundance than the stomach, duodenum, jejunum, and colon (P < 0.05). However, there were no differences in the pCaSR mRNA abundance observed among the stomach, duodenum, and jejunum (P > 0.05).
Figure 5.
Real-time RT-PCR analyses of pCaSR mRNA abundances in the kidney, lung, liver, stomach, duodenum, jejunum, ileum, and colon in the weaned piglets. Results were normalized with GAPDH as a house keeping gene and presented as mean ± SEM (n = 6). Bars show different letters differ (P < 0.05).
Western Blotting Analyses of pCaSR Protein Abundance in Different Tissues
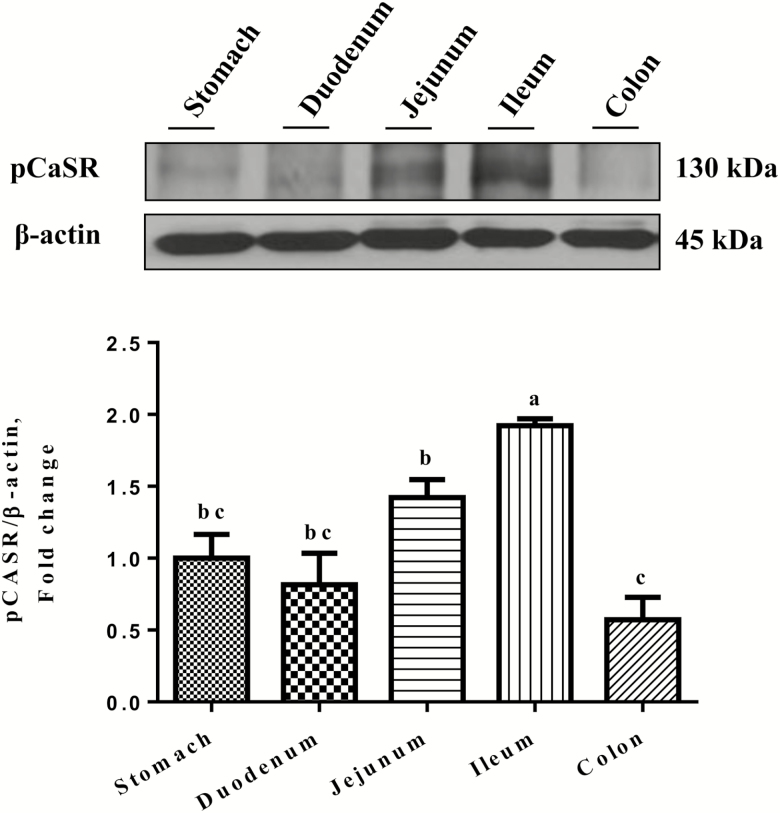
As shown in Figure 6, Western blotting analyses showed the presence of a 130-kDa pCaSR protein band in the tissue homogenates in the stomach, duodenum, jejunum, ileum, and colon in weaned piglets. The ileum had the highest level of the pCaSR protein when compared with the stomach, duodenum, jejunum, and colon (P < 0.05). There were no differences in pCaSR protein abundance among the stomach, duodenum, and jejunum (P > 0.05). The colon had the lowest level of the pCaSR protein, although there were no differences observed among the stomach, duodenum, and colon (P > 0.05).
Figure 6.
Western blotting analyses of pCaSR protein abundances in the stomach, duodenum, jejunum, ileum, and colon in the weaned piglets. (A) Representative bands of pCaSR (130 kDa) and β-actin (45 kDa). (B) Densitometric analyses of Western blotting bands. Data were normalized with β-actin as the housekeeping protein and presented as mean ± SEM (n = 3). Bars show different letters differ (P < 0.05).
Immunolocalization of the pCaSR Protein in Different Tissues
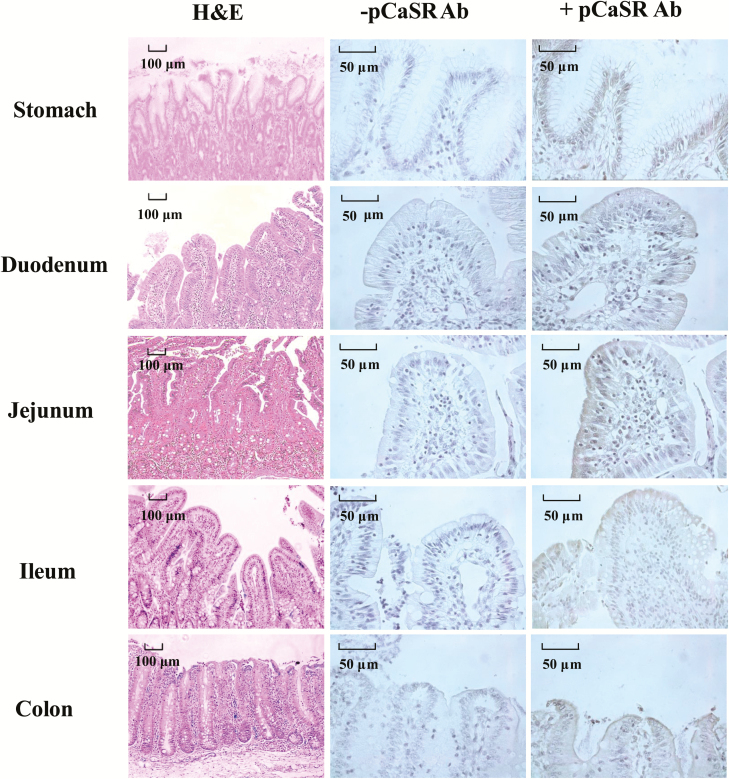
To further visually reveal pCaSR localization in the stomach, duodenum, jejunum, ileum, and colon in weaned piglets, immunohistochemical staining was performed. As shown in Figure 7, pCaSR protein expression was observed along the gastrointestinal tract. The pCaSR staining was localized predominantly on the simple columnar epithelium of stomach while no pCaSR staining was observed in the smooth muscle layers. There was pCaSR immunostaining detected on the surface of epithelial cells in the duodenum, jejunum, ileum, and colon (Figure 7). The CaSR staining was not observed, when the primary antibody was excluded in the negative control staining (Figure 7).
Figure 7.
Haemotoxylin and Eosin (H&E) staining and immunohistochemical staining of pCaSR protein in the stomach, duodenum, jejunum, ileum, and colon in the weaned piglets. Positive staining for pCaSR was indicated by brown deposits. −pCaSR represented negative control which the primary antibody was omitted from the staining procedure. Scale bar (black) for H&E staining represents 100 μm, and for immunohistochemical staining is 50 μm.
DISCUSSION
Weaning pigs are often associated with diarrhea, abnormal gut development, and impaired nutrient utilization (Liu et al., 2013). Newborn piglets receive most of their nutrients from sow’s milk that contains a high concentration of casein, spermine, spermidine, and Ca2+ (Cheng et al., 2006). These compounds allow for stimulation of pCaSR to maintain gut homeostasis. However, upon weaning of nursing pigs (transition from milk to solid feed), the nutrient source for maintaining gut tissue growth will be the solid feed with less pCaSR agonists such as spermidine and Ca2+. Ca2+ is required for development and maintenance of stable tight junction between epithelial cells (Galli et al., 1976; Martinez-Palomo et al., 1980). Activation of CaSR increased renal claudin-14 expression (Dimke et al., 2013) and improved tight junction proteins in the renal epithelial cell membrane (Jouret et al., 2013). Thus, it can be hypothesized that administering an effective amount of pCaSR modulators may improve nutrient absorption and gut barrier function in pigs, especially under challenging conditions. Although human CaSR has been intensively studied (Santos-Hernandez et al., 2018; Zhang and Mine, 2018), there is limited information about pCaSR. The present study was to investigate some basic structural features of the pCaSR gene and protein and their homology with the CaSR of other species.
Human CaSR gene spans ~103-kb (Yun et al., 2007) and the whole gene length of pCaSR is also about 104 kb. Alignment results showed a high degree of sequence homology between pCaSR and the CaSR of other mammalian species. This high existence of a homologous form of pCaSR suggests that the receptor is developed in some phylogenetic ancestor common to mammals. Meanwhile, like the CaSR of other species such as birds and mammals (Diaz et al., 1997; Hu and Spiegel, 2007; Alfadda et al., 2014; Hendy and Canaff, 2016), the putative protein structure of pCaSR shared similar protein structural features with them. Hence, pCaSR shares high resemblance with human CaSR regarding protein structure, which can be explained by the fact that pig physiology is similar to that of humans (Odle et al., 2014). All these demonstrate that the molecular characterization of pCaSR is highly similar to the other mammalian CaSR.
Even though the mRNA and protein sequences of pCaSR and human CaSR share some conservative regions, some variances were found within the signal peptide, ECD, TMDs and ICD of pCaSR, compared with human CaSR. According to a previous study, the signal peptide is predicted to target the nascent polypeptide chain of the CaSR preprotein and transfer it into the endoplasmic reticulum, so a functional difference may occur between pCaSR signal peptide and the one for human (Pidasheva et al., 2005). The ligands binding sites of pCaSR and the related activation may also be different from human CaSR because the majority of CaSR ligands binding sites are located in ECD (Brennan et al., 2013; Geng et al., 2016). Meanwhile, the early study showed that some key residues within TMD are vital for the ligands binding, and variances at some critical residues can reduce the effects of both NPS 2143 and NPS R-568 (Miedlich et al., 2004). Thus, this implies that when interacting with an antagonist or an agonist, the related effects may be different between human CaSR and pCaSR. Moreover, the ICD contains interaction sites for transduction coupling to multiple signaling pathways (Zhang et al., 2016), so the intracellular signaling pathways triggered by the activation of pCaSR could also vary from other species. CaSR can be activated by Ca2+, AAs, peptides, and specific pharmacological agonists in humans (Chattopadhyay, 1998; Ohsu et al., 2010). However, there may be other potential CaSR ligands and differences in response to ligands between human CaSR and pCaSR (Haid et al., 2012). Therefore, it would be critical to characterize the functions of pCaSR and its potential ligands, which might help to develop novel approaches to manipulate gut chemosensing through pCaSR for improving absorption, barrier function, and gut development during weaning or challenging conditions, reducing the dependence on the use of feed antibiotics in swine production.
In the present study, the expression pattern of pCaSR mRNA in different tissues of weaned piglets is consistent with previous studies (Cheng et al., 1999; Riccardi and Brown, 2010; Ward et al., 2012; Tang et al., 2016). It was reported that CaSR is widely distributed in tissues of the animal body and plays a crucial role in the animal’s physical activities (Cheng et al., 1999; Ward et al., 2012). CaSR is not only highly expressed in the kidney, bone tissues, thyroid, parathyroid and stomach but also in the pancreas, marrow, breasts, liver, and vascular smooth muscle (Tang et al., 2016). Our results also demonstrated that the mRNA level of pCaSR in the kidney is also the highest compared with other tissues and it is consistent with the high expression level of human CaSR in the kidney (Riccardi and Brown, 2010). These results suggest that CaSR is highly involved in mineral ion homeostasis in the kidney.
Western blotting analysis showed that the molecular weight of the pCaSR protein shares a similar size with the human CaSR (Bai et al., 1998; Desai et al., 2014). Moreover, our western blotting results also demonstrated that the pCaSR protein is widely expressed along the longitudinal axis of the digestive tract in weaned piglets. Considering the fact that the small intestine accounts for about 90% of overall calcium absorption (Wasserman, 2004; Breves, 2007), and the stomach or the large intestine can take up 10% of the total amount of absorbed calcium (Barger-Lux et al., 1989; Metzler-Zebeli et al., 2010; Gonzalez-Vega et al., 2014), this could explain the reason why a high expression level of pCaSR was verified in pig gastrointestinal tract, especially in small intestine.
Higher pCaSR protein abundance was found in the ileum of weaned piglets, which is consistent with the pCaSR mRNA abundance observed in the ileum. One potential reason is that the absorption of ingested calcium mainly occurs in the ileum. For instance, in the pig, over 60% of calcium absorption occurs in the ileum, >20% in the jejunum, and <10% in the duodenum, respectively (Partridge, 1978). In rats and dogs, the related values in the ileum, jejunum, and duodenum shared a similar pattern (Cramer, 1959; Ckam, 1965). Hence, the distribution pattern of pCaSR in pig gastrointestinal tract is consistent with the relative contribution of the different segments of the intestinal tract to overall calcium absorption. According to our immunohistochemistry results, it showed that positive staining in the apical membranes of both small intestine and colon, suggesting pCaSR is mainly expressed in the apical membranes of pig villus cells. An early study mirrored this result given that it claimed that CaSR appeared on the brush border of the intestine (Garg et al., 2013). However, it is difficult to determine which specific cells express pCaSR by using immunohistochemical staining.
The intestinal epithelium is lined with a single layer of epithelial cells including polarized absorptive enterocytes, goblet cells, enteroendocrine cells (EECs), and paneth cells, and absorptive enterocytes constitute the majority of the cells lining the villus (Cheng and Leblond, 1974; Hampson, 1986; Liou et al., 2011; Okumura and Takeda, 2017). CaSR has been widely detected in a variety of intestinal cells like EECs, but not including absorptive enterocytes (Chattopadhyay, 1998; Hira et al., 2008; Ohsu et al., 2010). However, some taste receptors were found in both absorptive enterocytes and EECs (Mace et al., 2007). Moreover, nutrients are detected by both absorptive enterocytes and EECs (Shirazi-Beechey et al., 2011) and CaSR has been found in enterocyte cell lines such as Caco-2 and HT-29 (Mine and Zhang, 2015a, b; Zhang et al., 2015). Thus, pCaSR could be expressed in the porcine native enterocytes and commercial porcine enterocyte cell lines. Further research is needed to confirm the expression of CaSR in the different types of epithelial cells isolated from weaned piglets.
CONCLUSION
The present study suggests that the AA sequence of pCaSR has variances when compared with the AA sequence of CaSR in other mammals, which might lead to its different functions with human CaSR. The pCaSR is widely expressed in the kidney, lung, liver, stomach, duodenum, jejunum, ileum, and colon in weaned piglets. Higher pCaSR mRNA and protein abundances were found in the ileum of weaned piglets, suggesting the nutrient sensing roles of pCaSR in the ileum. Further research is needed to confirm the expression of CaSR in the different types of epithelial cells isolated from weaned piglet’s intestine and characterize the functions of pCaSR, its potential ligands and cell signaling pathways related to CaSR activation in EECs and potentially in enterocytes.
Conflict of interest statement. None declared.
Footnotes
This work was supported by Natural Sciences and Engineering Council of Canada (NSERC) Discovery Grants (C. Yang: RGPIN-2017-04967), the University of Manitoba Start-Up Grant, and the Graduate Enhancement of Tri-Council Stipends (GETS) program at the University of Manitoba. The authors are grateful to Yulian Niu at the University of Manitoba histology lab for immunohistochemical analyses.
LITERATURE CITED
- Alfadda T. I., Saleh A. M., Houillier P., and Geibel J. P.. . 2014. Calcium-sensing receptor 20 years later. Am. J. Physiol. Cell Physiol. 307:C221–C231. doi: 10.1152/ajpcell.00139.2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aluko K., Velayudhan D. E., Khafipour E., Fang L., and Nyachoti M.. . 2017. Effect of chicken egg anti-F4 antibodies on performance and diarrhea incidences in enterotoxigenic Escherichia coli K88+-challenged piglets. Anim. Nutr. 3:353–358. doi: 10.1016/j.aninu.2017.06.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bai M., Trivedi S., and Brown E. M.. . 1998. Dimerization of the extracellular calcium-sensing receptor (car) on the cell surface of car-transfected HEK293 cells. J. Biol. Chem. 273:23605–23610. doi:10.1074/jbc.273.36.23605 [DOI] [PubMed] [Google Scholar]
- Barger-Lux M. J., Heaney R. P., and Recker R. R.. . 1989. Time course of calcium absorption in humans: evidence for a colonic component. Calcif. Tissue Int. 44:308–311. doi:10.1007/BF02556309 [DOI] [PubMed] [Google Scholar]
- Brennan S. C., Thiem U., Roth S., Aggarwal A., Fetahu I. S. h., Tennakoon S., Gomes A. R., Brandi M. L., Bruggeman F., Mentaverri R., . et al. 2013. Calcium sensing receptor signalling in physiology and cancer. Biochim. Biophys. Acta 1833:1732–1744. doi: 10.1016/j.bbamcr.2012.12.011 [DOI] [PubMed] [Google Scholar]
- Breves B. S. D. A. G. 2007. Mechanisms and regulation of calcium absorption from the gastrointestinal tract in pigs and ruminants: comparative aspects with special emphasis on hypocalcemia in dairy cows. Anim. Health. Res. Rev. 7:31–41. doi: 10.1017/S1466252307001144 [DOI] [PubMed] [Google Scholar]
- Burrin D., Stoll B., and Moore D.. . 2013. Digestive physiology of the pig symposium: intestinal bile acid sensing is linked to key endocrine and metabolic signaling pathways. J. Anim. Sci. 91:1991–2000. doi: 10.2527/jas.2013-6331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- CCAC. 2009. Guidelines on the care and use of farm animals in research, teaching and testing. Ottawa, ON: CCAC. [Google Scholar]
- Chattopadhyay N., Cheng I., Rogers K., Riccardi D., Hall A., Diaz R., Hebert S. C., Soybel D. I., and Brown E. M.. . 1998. Identification and localization of extracellular Ca2+-sensing receptor in rat intestine. Am. J. Physiol. 274(1 Pt 1):G122–G130. [DOI] [PubMed] [Google Scholar]
- Cheng S. X. 2016. Calcium-sensing receptor: a new target for therapy of diarrhea. World J. Gastroenterol. 22:2711–2724. doi: 10.3748/wjg.v22.i9.2711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng H., and Leblond C. P.. . 1974. Origin, differentiation and renewal of the four main epithelial cell types in the mouse small intestine. V. Unitarian theory of the origin of the four epithelial cell types. Am. J. Anat. 141:537–561. doi: 10.1002/aja.1001410407 [DOI] [PubMed] [Google Scholar]
- Cheng Z. B., Li D. F., Ge C. R., and Xing J. J.. . 2006. Polyamines in sow colostrum and milk at different stages of lactation. Anim. Sci. 82:95–99. doi: 10.1079/Asc20051 [DOI] [PubMed] [Google Scholar]
- Cheng I., Qureshi I., Chattopadhyay N., Qureshi A., Butters R. R., Hall A. E., Cima R. R., Rogers K. V., Hebert S. C., Geibel J. P., . et al. 1999. Expression of an extracellular calcium-sensing receptor in rat stomach. Gastroenterology 116:118–126. doi:10.1016/S0016-5085(99)70235-0 [DOI] [PubMed] [Google Scholar]
- Ckam C. F. 1965. Sites of calcium absorption and the calcium concentration of gut contents in the dog. Can. J. Physiol. Pharmacol. 43:75–78. doi:10.1152/ajplegacy.1968.214.4.814 [DOI] [PubMed] [Google Scholar]
- Cramer C. F., and Copp D. H.. . 1959. Progress and rate of absorption of radiostrontium through intestinal tracts of rats. Proc. Soc. Exp. Biol. Med. 102:514–517.doi:10.3181/00379727-102-25301 [DOI] [PubMed] [Google Scholar]
- Desai A. J., Roberts D. J., Richards G. O., and Skerry T. M.. . 2014. Role of receptor activity modifying protein 1 in function of the calcium sensing receptor in the human TT thyroid carcinoma cell line. PLoS One 9:e85237. doi: 10.1371/journal.pone.0085237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diaz R., Hurwitz S., Chattopadhyay N., Pines M., Yang Y., Kifor O., Einat M. S., Butters R., Hebert S. C., and Brown E. M.. . 1997. Cloning, expression, and tissue localization of the calcium-sensing receptor in chicken (Gallus domesticus). Am. J. Physiol. 273(3 Pt 2):R1008–R1016. doi: 10.1152/ajpregu.1997.273.3.R1008 [DOI] [PubMed] [Google Scholar]
- Dimke H., Desai P., Borovac J., Lau A., Pan W., and Alexander R. T.. . 2013. Activation of the Ca2+-sensing receptor increases renal claudin-14 expression and urinary Ca2+ excretion. Am. J. Physiol. Renal Physiol. 304:F761–F769. doi: 10.1152/ajprenal.00263.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galli P., Brenna A., Camilli de P., and Meldolesi J.. . 1976. Extracellular calcium and the organization of tight junctions in pancreatic acinar cells. Exp. Cell Res. 99:178–183.doi:10.1016/0014-4827(76)90694-7 [DOI] [PubMed] [Google Scholar]
- Garg M. K., Kalra S., and Mahalle N.. . 2013. The intestinal calcistat: determinant of clinical vitamin D deficiency. Indian J. Endocrinol. Metab. 17:780–783. doi: 10.4103/2230-8210.117187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng Y., Mosyak L., Kurinov I., Zuo H., Sturchler E., Cheng T. C., Subramanyam P., Brown A. P., Brennan S. C., Mun H. C., . et al. 2016. Structural mechanism of ligand activation in human calcium-sensing receptor. Elife. 5:e13662. doi: 10.7554/eLife.13662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Vega J. C., Walk C. L., Liu Y., and Stein H. H.. . 2014. The site of net absorption of Ca from the intestinal tract of growing pigs and effect of phytic acid, Ca level and Ca source on Ca digestibility. Arch. Anim. Nutr. 68:126–142. doi: 10.1080/1745039X.2014.892249 [DOI] [PubMed] [Google Scholar]
- Haid D. C., Jordan-Biegger C., Widmayer P., and Breer H.. . 2012. Receptors responsive to protein breakdown products in g-cells and d-cells of mouse, swine and human. Front. Physiol. 3:65. doi: 10.3389/fphys.2012.00065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hampson D. J. 1986. Alterations in piglet small intestinal structure at weaning. Res. Vet. Sci. 40:32–40. doi: 10.1016/S0034-5288(18)30482-X [DOI] [PubMed] [Google Scholar]
- Hassan Y. I., Lahaye L., Gong M. M., Peng J., Gong J., Liu S., Gay C. G., and Yang C.. . 2018. Innovative drugs, chemicals, and enzymes within the animal production chain. Vet. Res. 49:71. doi: 10.1186/s13567-018-0559-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hendy G. N., and Canaff L.. . 2016. Calcium-sensing receptor gene: regulation of expression. Front. Physiol. 7:394. doi: 10.3389/fphys.2016.00394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hira T., Nakajima S., Eto Y., and Hara H.. . 2008. Calcium-sensing receptor mediates phenylalanine-induced cholecystokinin secretion in enteroendocrine STC-1 cells. FEBS J. 275:4620–4626. doi: 10.1111/j.1742-4658.2008.06604.x [DOI] [PubMed] [Google Scholar]
- Hu J., and Spiegel A. M.. . 2007. Structure and function of the human calcium-sensing receptor: insights from natural and engineered mutations and allosteric modulators. J. Cell. Mol. Med. 11:908–922. doi: 10.1111/j.1582-4934.2007.00096.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ipharraguerre I. R. 2013. Digestive physiology of the pig symposium: gut chemosensing: integrating nutrition, gut function, and metabolism in pigs. J. Anim. Sci. 91:1929–1931. doi: 10.2527/jas.2013-6483 [DOI] [PubMed] [Google Scholar]
- Jouret F., Wu J., Hull M., Rajendran V., Mayr B., Schöfl C., Geibel J., and Caplan M. J.. . 2013. Activation of the Ca2+-sensing receptor induces deposition of tight junction components to the epithelial cell plasma membrane. J. Cell Sci. 126(Pt 22):5132–5142. doi: 10.1242/jcs.127555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaji I., Akiba Y., and Kaunitz J. D.. . 2013. Digestive physiology of the pig symposium: involvement of gut chemosensing in the regulation of mucosal barrier function and defense mechanisms. J. Anim. Sci. 91:1957–1962. doi: 10.2527/jas.2012-5941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liou A. P., Sei Y., Zhao X., Feng J., Lu X., Thomas C., Pechhold S., Raybould H. E., and Wank S. A.. . 2011. The extracellular calcium-sensing receptor is required for cholecystokinin secretion in response to L-phenylalanine in acutely isolated intestinal I cells. Am. J. Physiol. Gastrointest. Liver Physiol. 300:G538–G546. doi: 10.1152/ajpgi.00342.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Ipharraguerre I. R., and Pettigrew J. E.. . 2013. Digestive physiology of the pig symposium: potential applications of knowledge of gut chemosensing in pig production. J. Anim. Sci. 91(5):1982–1990. doi:10.2527/jas.2012–6193 [DOI] [PubMed] [Google Scholar]
- Liu W., Xie Y., Ma J., Luo X., Nie P., Zuo Z., Lahrmann U., Zhao Q., Zheng Y., Zhao Y., . et al. 2015. IBS: an illustrator for the presentation and visualization of biological sequences. Bioinformatics. 31:3359–3361. doi: 10.1093/bioinformatics/btv362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., and Schmittgen T. D.. . 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 25:402–408. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- Mace O. J., Affleck J., Patel N., and Kellett G. L.. . 2007. Sweet taste receptors in rat small intestine stimulate glucose absorption through apical GLUT2. J. Physiol. 582(Pt 1):379–392. doi: 10.1113/jphysiol.2007.130906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mace O. J., and Marshall F.. . 2013. Digestive physiology of the pig symposium: gut chemosensing and the regulation of nutrient absorption and energy supply. J. Anim. Sci. 91:1932–1945. doi: 10.2527/jas.2012-5906 [DOI] [PubMed] [Google Scholar]
- Martinez-Palomo A., Meza I., Beaty G., and Cereijido M.. . 1980. Experimental modulation of occluding junctions in a cultured transporting epithelium. J. Cell Biol. 87(3 Pt 1):736–745. doi:10.1083/jcb.87.3.736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maruyama Y., Yasuda R., Kuroda M., and Eto Y.. . 2012. Kokumi substances, enhancers of basic tastes, induce responses in calcium-sensing receptor expressing taste cells. PLoS One. 7:e34489. doi: 10.1371/journal.pone.0034489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzler-Zebeli B. U., Hooda S., Mosenthin R., Gänzle M. G., and Zijlstra R. T.. . 2010. Bacterial fermentation affects net mineral flux in the large intestine of pigs fed diets with viscous and fermentable nonstarch polysaccharides. J. Anim. Sci. 88:3351–3362. doi: 10.2527/jas.2010-2906 [DOI] [PubMed] [Google Scholar]
- Miedlich S. U., Gama L., Seuwen K., Wolf R. M., and Breitwieser G. E.. . 2004. Homology modeling of the transmembrane domain of the human calcium sensing receptor and localization of an allosteric binding site. J. Biol. Chem. 279:7254–7263. doi: 10.1074/jbc.M307191200 [DOI] [PubMed] [Google Scholar]
- Mine Y., and Zhang H.. . 2015a. Anti-inflammatory effects of poly-L-lysine in intestinal mucosal system mediated by calcium-sensing receptor activation. J. Agric. Food Chem. 63:10437–10447. doi: 10.1021/acs.jafc.5b03812 [DOI] [PubMed] [Google Scholar]
- Mine Y., and Zhang H.. . 2015b. Calcium-sensing receptor (CASR)-mediated anti-inflammatory effects of L-amino acids in intestinal epithelial cells. J. Agric. Food Chem. 63:9987–9995. doi: 10.1021/acs.jafc.5b03749 [DOI] [PubMed] [Google Scholar]
- Odle J., Lin X., Jacobi S. K., Kim S. W., and Stahl C. H.. . 2014. The suckling piglet as an agrimedical model for the study of pediatric nutrition and metabolism. Annu. Rev. Anim. Biosci. 2:419–444. doi: 10.1146/annurev-animal-022513-114158 [DOI] [PubMed] [Google Scholar]
- Ohsu T., Amino Y., Nagasaki H., Yamanaka T., Takeshita S., Hatanaka T., Maruyama Y., Miyamura N., and Eto Y.. . 2010. Involvement of the calcium-sensing receptor in human taste perception. J. Biol. Chem. 285:1016–1022. doi: 10.1074/jbc.M109.029165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura R., and Takeda K.. . 2017. Roles of intestinal epithelial cells in the maintenance of gut homeostasis. Exp. Mol. Med. 49:e338. doi: 10.1038/emm.2017.20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen J. L., Cheng S. X., Ge Y., Sahay B., and Mohamadzadeh M.. . 2016. The role of the calcium-sensing receptor in gastrointestinal inflammation. Semin. Cell Dev. Biol. 49:44–51. doi: 10.1016/j.semcdb.2015.10.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partridge I. G. 1978. Studies on digestion and absorption in the intestines of growing pigs. 3. Net movements of mineral nutrients in the digestive tract. Br. J. Nutr. 39:527–537. doi:10.1079/BJN19780068 [DOI] [PubMed] [Google Scholar]
- Pidasheva S., Canaff L., Simonds W. F., Marx S. J., and Hendy G. N.. . 2005. Impaired cotranslational processing of the calcium-sensing receptor due to signal peptide missense mutations in familial hypocalciuric hypercalcemia. Hum. Mol. Genet. 14:1679–1690. doi: 10.1093/hmg/ddi176 [DOI] [PubMed] [Google Scholar]
- Reimann F., Tolhurst G., and Gribble F. M.. . 2012. G-protein-coupled receptors in intestinal chemosensation. Cell Metab. 15:421–431. doi: 10.1016/j.cmet.2011.12.019 [DOI] [PubMed] [Google Scholar]
- Riccardi D., and Brown E. M.. . 2010. Physiology and pathophysiology of the calcium-sensing receptor in the kidney. Am. J. Physiol. Renal Physiol. 298:F485–F499. doi: 10.1152/ajprenal.00608.2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santos-Hernandez M., Miralles B., Amigo L., and Recio I.. . 2018. Intestinal signaling of proteins and digestion-derived products relevant to satiety. J. Agric. Food Chem. 66:10123–10131. doi: 10.1021/acs.jafc.8b02355 [DOI] [PubMed] [Google Scholar]
- Shirazi-Beechey S. P., Moran A. W., Bravo D., and Al-Rammahi M.. . 2011. Nonruminant nutrition symposium: intestinal glucose sensing and regulation of glucose absorption: implications for swine nutrition. J. Anim. Sci. 89:1854–1862. doi: 10.2527/jas.2010-3695 [DOI] [PubMed] [Google Scholar]
- Song T., Peng J., Ren J., Wei H. K., and Peng J.. . 2015. Cloning and characterization of spliced variants of the porcine G protein coupled receptor 120. Biomed Res. Int. 2015:813816. doi: 10.1155/2015/813816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Symonds E. L., Peiris M., Page A. J., Chia B., Dogra H., Masding A., Galanakis V., Atiba M., Bulmer D., Young R. L., . et al. 2015. Mechanisms of activation of mouse and human enteroendocrine cells by nutrients. Gut 64:618–626. doi: 10.1136/gutjnl-2014-306834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., and Kumar S.. . 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30:2725–2729. doi: 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang L., Cheng C. Y., Sun X., Pedicone A. J., Mohamadzadeh M., and Cheng S. X.. . 2016. The extracellular calcium-sensing receptor in the intestine: evidence for regulation of colonic absorption, secretion, motility, and immunity. Front. Physiol. 7:245. doi: 10.3389/fphys.2016.00245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tedo G., Roura E., Ipharraguerre I., and X. Manteca. 2011. Gene expression of the L-amino acid–sensing receptor T1R1/T1R3 changes in gut tissues of pigs in response to dietary protein. J. Anim. Sci. 89:86. doi: 10.1016/j.bbrc.2011.02.016 [DOI] [Google Scholar]
- Van Boeckel T. P., Brower C., Gilbert M., Grenfell B. T., Levin S. A., Robinson T. P., Teillant A., and Laxminarayan R.. . 2015. Global trends in antimicrobial use in food animals. Proc. Natl. Acad. Sci. USA. 112:5649–5654. doi: 10.1073/pnas.1503141112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward B. K., Magno A. L., Walsh J. P., and Ratajczak T.. . 2012. The role of the calcium-sensing receptor in human disease. Clin. Biochem. 45:943–953. doi: 10.1016/j.clinbiochem.2012.03.034 [DOI] [PubMed] [Google Scholar]
- Wasserman R. H. 2004. Vitamin D and the dual processes of intestinal calcium absorption. J. Nutr. 134:3137–3139. doi: 10.1093/jn/134.11.3137 [DOI] [PubMed] [Google Scholar]
- Wu H. X., Wang C., Gregory K. J., Han G. W., Cho H. P., Xia Y., Niswender C. M., Katritch V., Meiler J., Cherezov V., . et al. 2014. Structure of a class C GPCR metabotropic glutamate receptor 1 bound to an allosteric modulator. Science. 344:58–64. doi: 10.1126/science.1249489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., Albin D. M., Wang Z., Stoll B., Lackeyram D., Swanson K. C., Yin Y., Tappenden K. A., Mine Y., Yada R. Y., . et al. 2011. Apical Na+-D-glucose cotransporter 1 (SGLT1) activity and protein abundance are expressed along the jejunal crypt-villus axis in the neonatal pig. Am. J. Physiol. Gastrointest. Liver Physiol. 300:G60–G70. doi: 10.1152/ajpgi.00208.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., Chowdhury M. A., Huo Y., and Gong J.. . 2015. Phytogenic compounds as alternatives to in-feed antibiotics: potentials and challenges in application. Pathogens 4:137–156. doi: 10.3390/pathogens4010137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang C., Huang T., Wu L., Li T., Wu G., and Yin Y.. . 2013. Molecular cloning and characterization of porcine calcium-sensing receptor (CaSR). Amino Acids. 45:598. doi:10.1007/s00726-013-1540-y [Google Scholar]
- Yang C., Yang X., and Fan M. Z.. . 2016a. Apical Na+-dependent neutral amino acid exchanger ASCT2 (ATB0) and mTOR-signaling components are expressed along the entire jejunal crypt-villus axis in young pigs fed a liquid milk replacer. Can. J. Anim. Sci. 97:19-29. doi: 10.1139/cjas-2016-0021 [DOI] [Google Scholar]
- Yang C., Yang X., Lackeyram D., Rideout T. C., Wang Z., Stoll B., Yin Y., Burrin D. G., and Fan M. Z.. . 2016b. Expression of apical Na+-L-glutamine co-transport activity, B0-system neutral amino acid co-transporter (B0AT1) and angiotensin-converting enzyme 2 along the jejunal crypt-villus axis in young pigs fed a liquid formula. Amino Acids. 48:1491–1508. doi: 10.1007/s00726-016-2210-7 [DOI] [PubMed] [Google Scholar]
- Yun F. H., Wong B. Y., Chase M., Shuen A. Y., Canaff L., Thongthai K., Siminovitch K., Hendy G. N., and Cole D. E.. . 2007. Genetic variation at the calcium-sensing receptor (CASR) locus: implications for clinical molecular diagnostics. Clin. Biochem. 40:551–561. doi: 10.1016/j.clinbiochem.2006.12.011 [DOI] [PubMed] [Google Scholar]
- Zhang H., Kovacs-Nolan J., Kodera T., Eto Y., and Mine Y.. . 2015. γ-glutamyl cysteine and γ-glutamyl valine inhibit TNF-α signaling in intestinal epithelial cells and reduce inflammation in a mouse model of colitis via allosteric activation of the calcium-sensing receptor. Biochim. Biophys. Acta. 1852:792–804. doi: 10.1016/j.bbadis.2014.12.023 [DOI] [PubMed] [Google Scholar]
- Zhang H., and Mine Y.. . 2018. Is calcium-sensing receptor a new molecular target toward improving gastrointestinal health? J. Agric. Food. Chem. 66:3995–3997. doi: 10.1021/acs.jafc.8b01150 [DOI] [PubMed] [Google Scholar]
- Zhang C., Zhang T., Zou J., Miller C. L., Gorkhali R., Yang J. Y., Schilmiller A., Wang S., Huang K., Brown E. M., . et al. 2016. Structural basis for regulation of human calcium-sensing receptor by magnesium ions and an unexpected tryptophan derivative co-agonist. Sci. Adv. 2:e1600241. doi: 10.1126/sciadv.1600241 [DOI] [PMC free article] [PubMed] [Google Scholar]