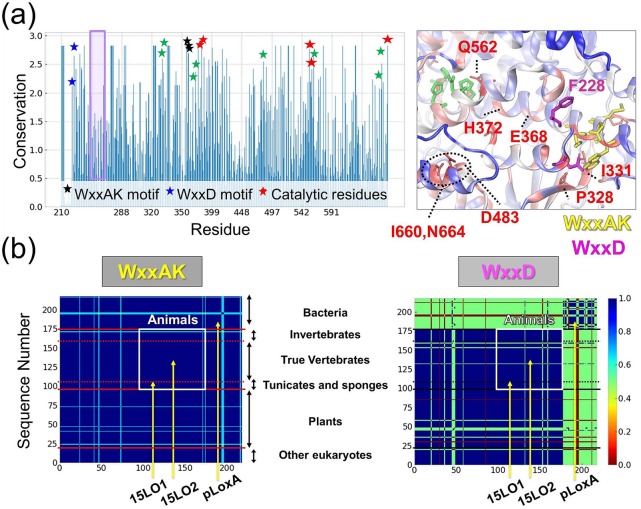
Figure 5.
Conservation of characteristic motifs present in LOXs. (a) Conservation propensity of LOX residues. Shannon entropy subtracted from maximum entropy is shown for the LOX catalytic domain (residues G217-Y671 in pLoxA) based on Pfam data. The highest values correspond to the most conserved residues. Stars show the WxxAK motif (black), the WxxD motif (blue), the catalytic residues (red), and the residues I331, P328, I660, N664, E368, H372, and D483 (green) with a relatively high level of conservation, displayed in the inset. The inset in the right panel displays conservation mapped on the 3D structure of pLoxA. Residues with the highest conservation are displayed in red, residues with a moderate level of conservation are displayed in white, and residues with a weak level of conservation are displayed in blue with the exception of the highly conserved WxxAK (yellow sticks) and WxxD (magenta) motifs. (b) Sequence identity matrices corresponding to the WxxAK (left) and WxxD (right) motifs obtained from 218 LOX sequences from all domains of life. High sequence similarity is in blue, intermediate sequence similarity is in yellow/green, and low sequence similarity is in red. The matrix generated for the entire LOX sequence in our previous study29 shows blocks evident for plants, animals, and bacteria that are indicated here. Residues from the WxxAK motif (left panel) are highly conserved in all species, whereas residues from the WxxD motif (right panel) exhibit a moderate level of conservation with species-specific differentiation with bacteria having dissimilar sequences to eukaryotes.