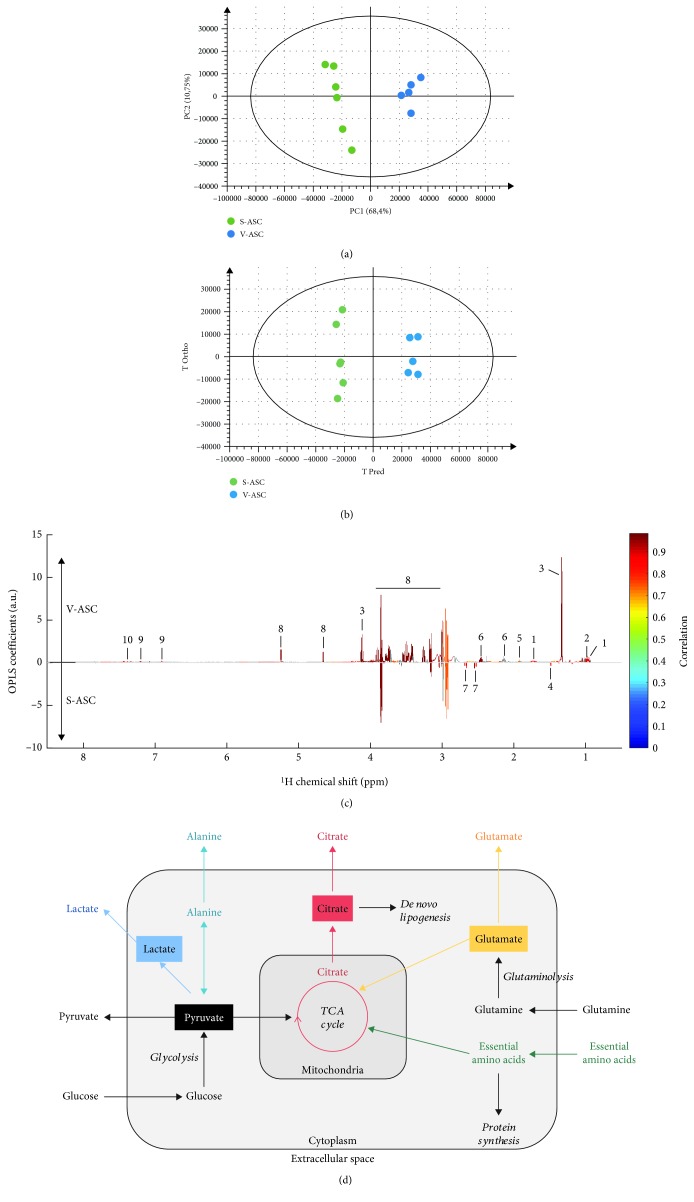
Figure 4.
Multivariate statistical analyses of the differences between S- and V-ASC exometabolomes. High-resolution NMR multivariate data analyses unveil significant differences in extracellular metabolite variations between S-ASC and V-ASC at 75% confluency and cultivated for additional 72 h in media containing 25 mM glucose, 4 mM glutamine, 1 mM pyruvate, and nonessential amino acids as determined by NMR absolute spectra bins after subtraction of the culture medium signal analysis performed on NMR spectra buckets without normalization to the cell number. At the time of supernatant collection, cells were at confluency. (a) Untargeted principal component analysis (PCA) readily evidences the cell type as a major origin of the dataset variance. Score plot of the PCA model (PC1 and PC2) (n = 11, R 2 = 0.962, and Q 2 = 0.843 on 5 principal components). (b, c) Supervised multivariate data analysis (O-PLS-DA) optimizes the discrimination between both cell types after 72 h of culture. The strong discrimination of the multivariate model is shown by the high values of goodness-of-fit model parameters R 2 and Q 2 (R 2(X) = 0.796, R 2(Y) = 0.991, and Q 2 = 0.969). The discrimination robustness was validated by resampling 1000 times the model under the null hypothesis (data not shown), and the analysis of variance (CV-ANOVA) of the model led to a p value of 1.20 × 10−4. (b) Score plot of the (1 + 1) O-PLS-DA model discriminating S-ASC (in green) and V-ASC (in blue). (c) O-PLS-DA loading plot after SRV analysis and Benjamini–Hochberg multiple testing correction. The O-PLS-DA loadings reveal the influential metabolite variations on the cell type discrimination (V-ASC upper panel and S-ASC lower panel). The loading plot was complemented by color-coded correlation [26] indicating statistically significant signals. Highlighted candidate biomarkers are (1) leucine, (2) valine, (3) lactate, (4) alanine, (5) acetate, (6) glutamine, (7) citrate, (8) glucose, (9) tyrosine, and (10) phenylalanine. (d) Simplified and nonquantitative representation of the main carbon metabolic pathways in actively dividing cells. Anabolic and catabolic pathways are represented in italics in relation to the metabolite secretome. Colors identify the metabolic pathways analyzed in this study. TCA: tricarboxylic acid cycle; EAA: essential amino acids.