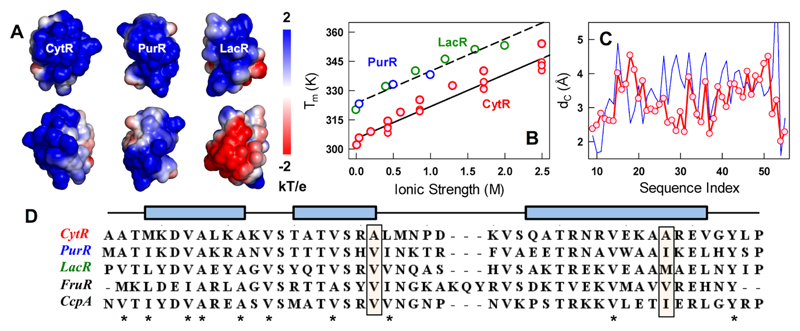
Figure 1.
Origins of disorder in the LacI DBD family. (A) Electrostatic potential maps of CytR, PurR and LacR (298 K, 43 mM ionic strength, pH 7.0) highlighting the electrostatic frustration at the DNA-binding face (top row) and the opposite face (bottom row). Note that the DNA-bound conformation of CytR is used for the calculation. (B) Observed changes in melting temperatures from far-UV CD experiments as a function of ionic strength for CytR (red), PurR (blue) and LacR (green). (C) Predicted differences in coupling distances as a function of the residue index for the DNA-bound CytR (red) and apo-PurR (blue). The PDB numbering of CytR starting from 9 is used as a reference (PDB id: 2L8N/2LCV). (D) Multiple sequence alignment of LacI family members with known structures. Stars represent the highly conserved apolar positions across all sequences. Rectangular filled boxes highlight positions 29 and 48 in CytR with small apolar side-chains contrasting with the rest of the sequences that display bulkier side-chains. Blue rectangular boxes above the sequences show the helical boundaries.