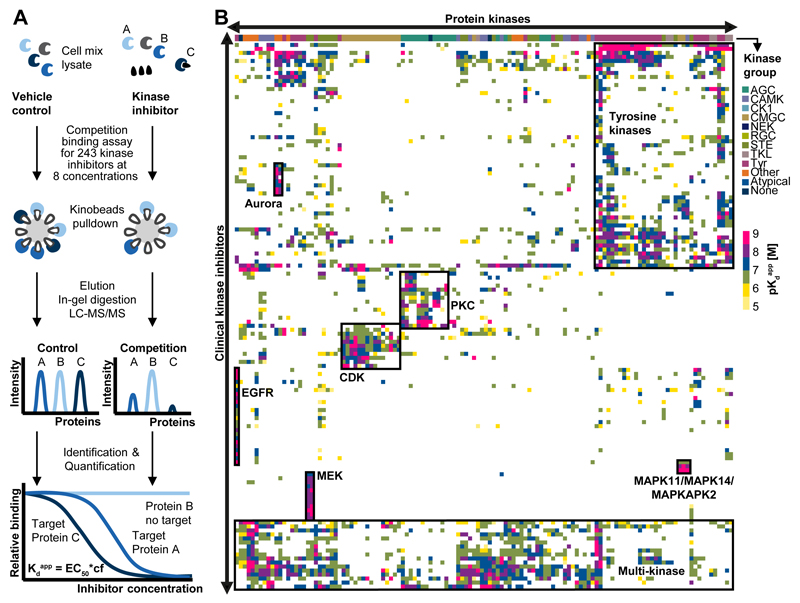
Figure 1.
The ‘druggable kinome’. (A) Schematic representation of the chemical proteomic workflow used to profile drug-protein interactions. Cell lysates were separately equilibrated with vehicle or increasing concentrations of each drug. Kinobeads were used to enrich kinases and other proteins from each lysate. Proteins were eluted from the beads, digested with trypsin, identified by liquid chromatography tandem mass spectrometry (LC-MS/MS) and quantified using the MS intensity of the identified peptides. Nonlinear regression determined the effective drug concentration at which half of the target protein was competed (EC50). EC50 values were converted into apparent dissociation constants (Kdapp) of each drug-target interaction by correcting for the depletion of a target protein from the lysate by Kinobeads (correction factor, cf). This workflow enabled the simultaneous measurement of interaction of one drug with hundreds of proteins in a single experiment. (B) Hierarchical clustering of kinase targets against clinical kinase drugs provided an overview of the ‘druggable’ kinome (color code depicts the Kdapp of drug-target interactions). Boxed regions represent groups of kinases and inhibitors thereof that ranged from highly selective (e.g., EGFR or MEK inhibitors) to relatively unselective interactions (e.g., tyrosine kinase and multi-kinase inhibitors). Further details are provided in Figures S1-2, Table S2-3 and the Supplementary Materials.