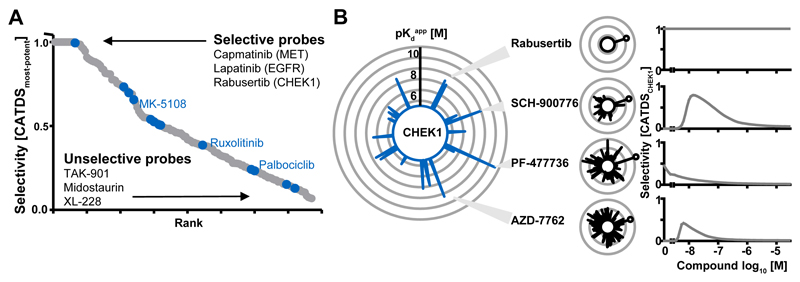
Figure 2.
Selectivity of kinase inhibitors. The collective drug-protein interaction data enabled the definition of a new selectivity metric (Concentration and Target Dependent Selectivity, CATDS). CATDS measures the reduction in binding of a target protein to Kinobeads at a specified concentration relative to the summed reduction of all protein targets of the compound at the same concentration. (A) Rank-plot of kinase inhibitors according to CATDSmost-potent (most potent compound target at the respective Kdapp) showing that Lapatinib, Capmatinib and Rabusertib are highly selective inhibitors; whilst TAK-901, Midostaurin and XL-228 are not. Compounds previously designated as ‘chemical probes’ are shown in blue but are not necessarily selective. (B) The large radar plot shows all CHEK1 inhibitors (each spoke is a drug and the length of the spoke is indicative of binding affinity). The smaller plots depict the number and potency of targets for Rabusertib, SCH-900776, PF-477736 and AZD-7762. The plot to the right shows that the selectivity of each compound (CATDSCHEK1 at its Kdapp) is a function of drug concentration. AZD-7762 is a potent CHEK1 inhibitor, but is not selective at any concentration. PF-477736 and SCH-900776 are selective at lower doses and Rabusertib is selective at all doses as no other targets beside CHEK1 were observed in this screen. Further details are provided in Figures S3-4 and the Supplementary Materials.