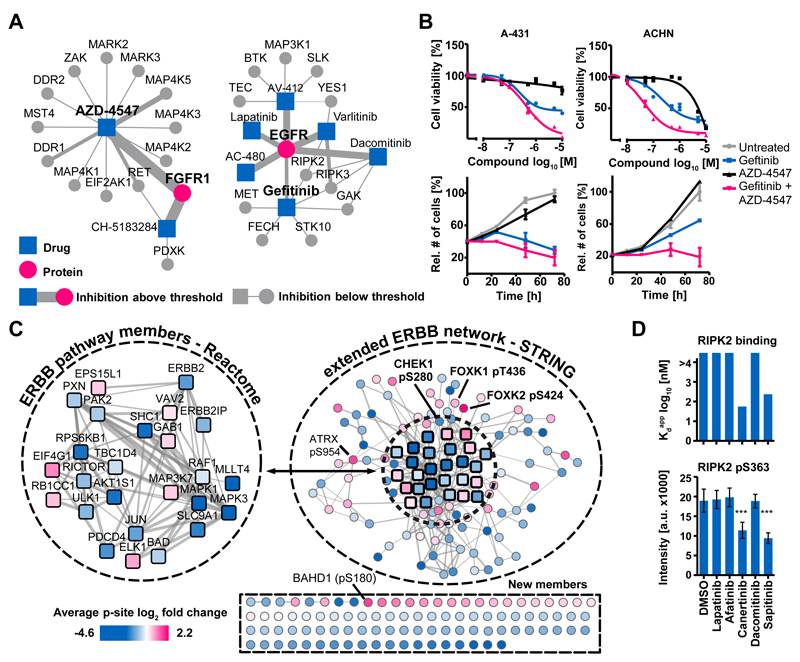
Figure 4.
From target to pathway engagement. (A) Visualization of protein-drug interactions in ProteomicsDB. Each node is a drug or a target and the size of each edge is proportional to the pKdapp of the interaction. Exploration of these networks can identify rational drug combinations to e.g., overcome drug resistance. (B) A-431 and ACHN cells that are partially sensitive to the EGFR inhibitor Gefitinib were treated with Gefitinib, the FGFR inhibitor AZD-4547, or a combination of both. Error bars depict standard error of the mean of technical triplicates. Cell viability and proliferation assays showed that the combination of the drugs was more effective than single compounds. (C) Quantitative phosphoproteomics was used to measure pathway engagement and identify common effects exerted by five kinase inhibitors (Lapatinib, Afatinib, Canertinib, Dacomitinib and Sapitinib) in the ERBB2-driven breast cancer cell line BT-474. Numerous phosphorylation sites mapped to known ERBB pathway members (Reactome) and further proteins were associated with the ERBB network using STRING. Many further phosphorylation sites were consistently and statistically significantly regulated by the drugs (p<0.01; scale: average log2 fold change across all inhibitors). These may represent novel functional effector proteins or pathway biomarkers of EGFR signaling and drug response. (D) Conversely, using the phosphorylation status of pS363 of RIPK2 is not reliable response marker for EGFR drugs because phosphorylation abundance of this site was only reduced by two from five of the EGFR inhibitors (bottom panel, error bars depict standard deviation) and instead only responded to inhibitors that are also RIPK2 inhibitors (top panel). Further details are provided in Figure S8 and the Supplementary Materials.