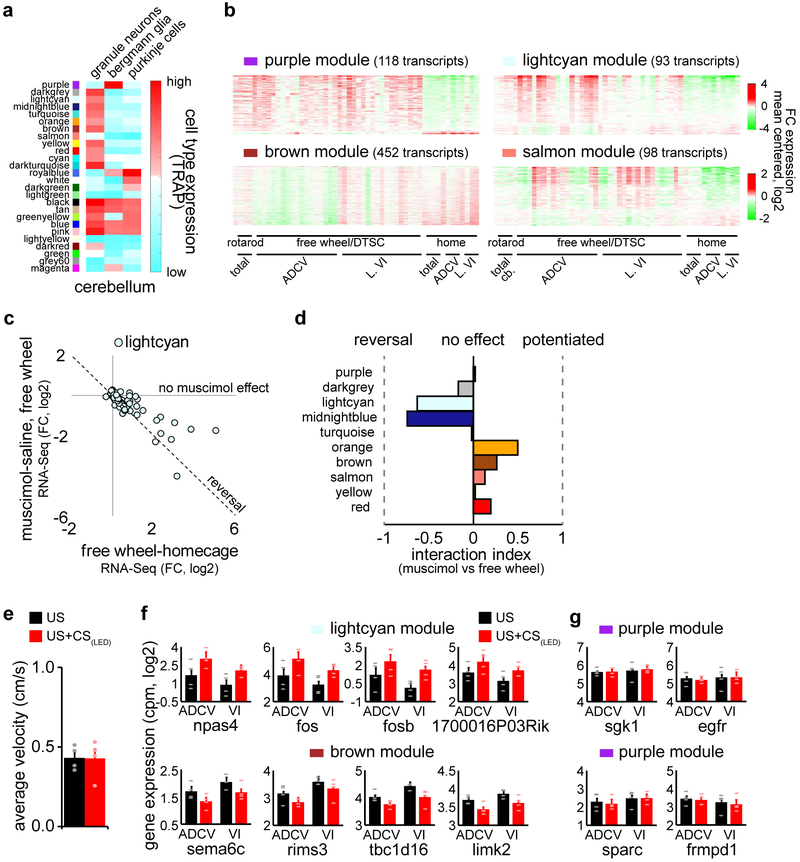
Extended Data Fig. 5. Identification of cell type-enriched and sensorimotor experience-dependent gene modules in the cerebellum in mice.
(a) Gene modules identified in Fig. 3b were intersected with cerebellar cell type-specific TRAP-Seq data and analyzed as in Fig. 3c. (b) Heatmaps of gene module expression induced by sensorimotor stimulation as in Fig. 3a (n=52 samples, log2 mean centered). (c) Comparison of the log2 fold change in gene expression induced by free wheel locomotion compared to homecage control and the log2 fold change in gene expression upon treatment with the GABA(A) receptor agonist muscimol compared to saline control during free wheel locomotion in the ADCV of mice. Genes in the lightcyan module induced by locomotion that were restored to homecage control expression levels upon silencing of the ADCV cortical activity with muscimol (reversal) fall on the dotted line. (d) Sensorimotor stimulation-dependent gene modules analyzed as in (c). (e) The average velocity of mice subjected to delay tactile startle conditioning (US+CS) or control (US) condition during inter-trial intervals (n=4 mice). (f, g) Gene expression in the ADCV or lobule VI of mice subjected to delay tactile startle conditioning (US+CS) or control (US) condition (n=4 mice). In all panels, data show mean and error bars denote standard error