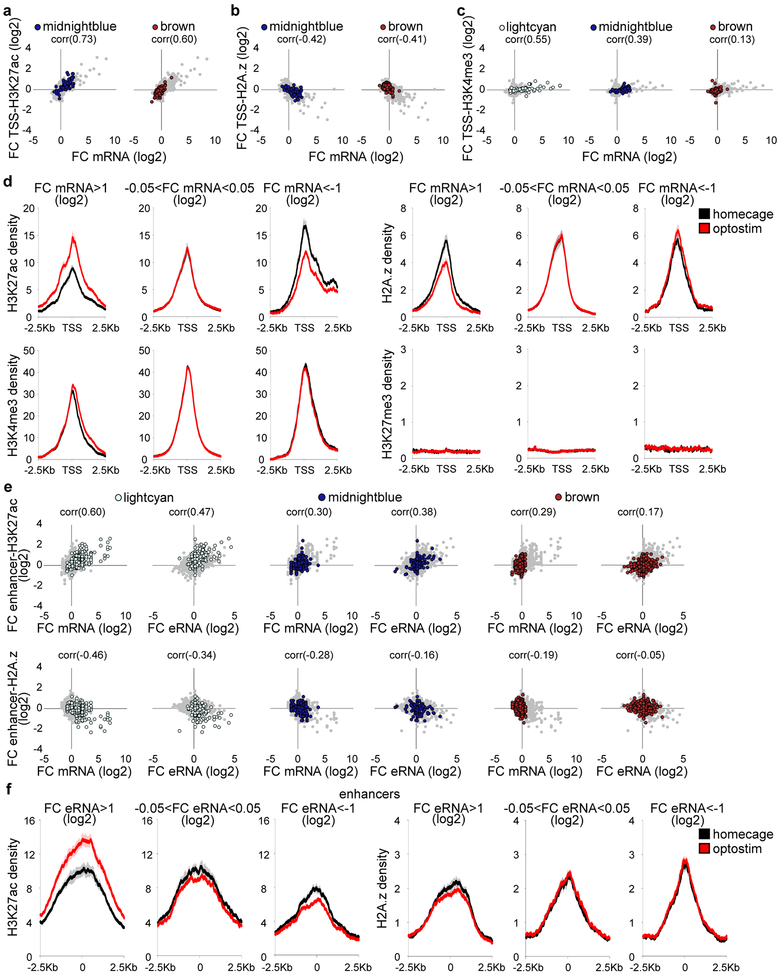
Extended Data Fig. 8. Optostimulation of the CS pathway regulates histone modification and histone variant abundance at gene promoters.
and enhancers (a-c) Comparison of the log2 fold change in gene expression and the log2 fold change in H3K27ac (a), H2A.z (b), or H3K4me3 (c) read density at the TSSs of granule neuron-enriched gene modules upon optostimulation of granule neurons in the ADCV in mice (n=85,355 transcripts for midnightblue, brown). The Pearson correlation coefficient (corr) is shown. (d) The profile of the histone marks H3K27ac, H2A.z, H3K4me3, and H3K27me3 surrounding the TSSs of genes whose expression was upregulated (left, mRNA fold change>1, log2), not changed (middle, −0.05<mRNA fold change<0.05, log2), or downregulated (right, mRNA fold change<−1, log2) upon optostimulation of granule neurons (n=2,3,2,2 biological replicates for H3K27ac,H2A.z,H3K4me3,H3K27me3). (e, f) Comparison of the log2 fold change in mRNA or eRNA and the log2 fold change in H3K27ac (top) or H2A.z (bottom) levels at the enhancers of granule neuron-enriched CS-regulated gene modules (e, n=266,256,1510 enhancers for lightcyan, midnightblue, brown). The Pearson correlation coefficient (corr) is shown. The profile of H3K27ac and H2A.z surrounding enhancers with eRNA levels that were upregulated (left, mRNA fold change>1, log2), not changed (middle, −0.05<mRNA fold change<0.05, log2), or downregulated (right, mRNA fold change<−1, log2) upon optostimulation of granule neurons (f, n=2,3 biological replicates for H3K27ac,H2A.z). In all panels, data show mean.