Fig. 6.
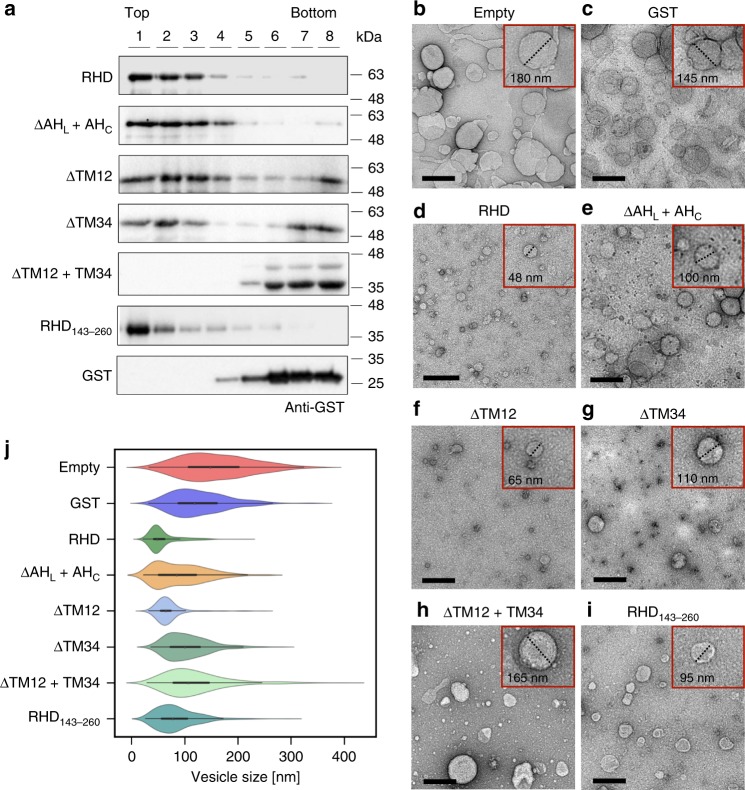
RHD structure determines in vitro membrane binding and liposome remodeling activity. a Liposome co-flotation assay to evaluate membrane-binding properties of FAM134B-RHD and various deletion mutants (see the section “Methods”). Purified protein samples were incubated with liposomes for 2 h at 37 °C and subjected to flotation on a sucrose cushion (top to bottom, 1–8) followed by SDS–PAGE and western blotting with anti-GST antibody. b–i Representative nsTEM micrographs of remodeled proteoliposomes (scale bars, 200 nm). Empty liposomes (b) were remodeled by incubation after addition of purified (c) GST, (d) wild-type RHD, (e) ΔAHL + AHC, (f) ΔTM12, (g) ΔTM34, (h) ΔTM12 + TM34, and (i) RHD143–260 for 18 h at 22 °C. Insets (red squares); magnified micrographs showing examples of representative proteoliposomes with diameters measured (dotted lines) using ImageJ. j Violin plots show the measured proteoliposome size-distributions (n = 300 each) from nsTEM images. Violins shows a central boxplot (median with interquartile range, black lines) along with mirrored histograms on either sides (colored)