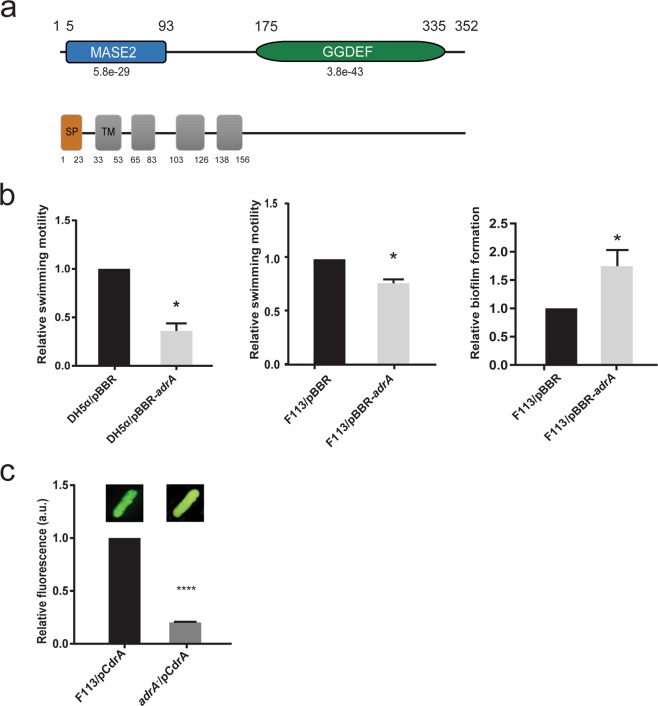
Figure 1.
AdrA encoded by PSF113_1982 in Pseudomonas fluorescens F113 is a DGC. (a) Predicted domain organization for AdrA protein (352 aa) from P. fluorescens F113 according to HMMER using profile hidden Markov models and Pfam database. Domains are indicated in each block. Domains (MASE2 and GGDEF) and their individual E-values are shown above. Signal peptide (SP, brown rectangle) and transmembrane domains (TM, grey rectangles) are shown below. Numbers indicate the start and end aa positions covered by each domain or feature. (b) Relative swimming motility in DH5α and P. fluorescens F113 and relative biofilm formation in P. fluorescens F113 in AdrA overexpression experiments. pBBRMCS-5 vector harbouring adrA from P. fluorescens F113 was used for overexpression experiments in both strains. The empty pBBRMCS-5 vector was used as control. Mean ± SD of three replicates are shown. Statistically significant difference (p < 0.05) are denoted by asterisks. (c) AdrA participates in the synthesis of the second messenger c-di-GMP. Streaks on LB medium of P. fluorescens F113 and its adrA mutant harbouring the gfp-based pCdra biosensor for c-di-GMP. Pictures were obtained in a Leika binocular microscope with a GFP filter set and 50 miliseconds of exposition time. Intracelullar levels of c-di-GMP were measured as fluorescence emission in the pCdra-containing strains. Mean ± SD of five analyzed extracts per strain are represented. Asterisks denote statistical significance of the data (****p < 0.0001).