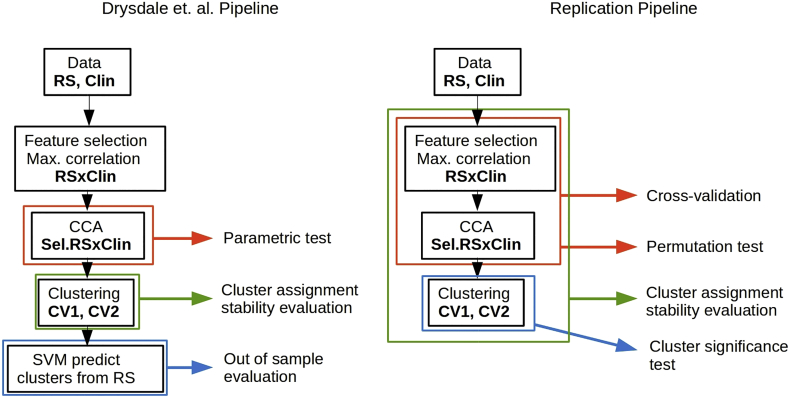
Fig. 1.
A scheme of the pipeline used in the original study and our pipeline. Data: in the original study, 220 depressed subjects have been analyzed as a part of a “cluster discovery” set and an additional 92 subjects were used as evaluation set. The clinical data (Clin) consisted of 17 HAM-D items. We have used 187 subjects with depression, anxiety disorder or depression-anxiety comorbidity. The clinical data consisted of 17 IDS items that best-matched the HAM-D item used in the original study. After preprocessing of fMRI data (RS), a correlation matrix between selected regions was created, resulting in ~35,000 features. A small subset of features (178 in the original study and 150 in our study) were selected based on their correlation with clinical symptoms (Sel.RS). Then, CCA was performed using these selected features and clinical symptoms. In the original study, a parametric test was used to the established statistical significance of CCA without taking a previous feature selection into an account. Hierarchical clustering was performed on first two resting state connectivity canonical variates (CV1, CV2). We have included an additional test, to test if the data cluster more than what is expected from data sampled from a Gaussian distribution. Stability of cluster assignment was evaluated in the original study by resampling of CV1 and CV2, We have extended the resampling stability evaluation to feature selection (in addition to the CCA procedures). Out of sample evaluation: in the original study, an additional 92 subjects were assigned to clusters according to a SVM model and clinical profiles of these clusters were compared to clinical profiles of clusters obtained in the cluster discovery set. We have evaluated the reproducibility of canonical correlations directly, using 10-fold cross-validation.