Abstract
Background
Subject C135 is one of the members of the Sydney Blood Bank Cohort, infected in 1981 through transfusion with attenuated nef/3′ long terminal repeat (LTR)-deleted HIV-1, and has maintained undetectable plasma viral load and steady CD4 cell count, in the absence of therapy. Uniquely, C135 combines five factors separately associated with control of viraemia: nef/LTR-deleted HIV-1, HLA-B57, HLA-DR13, heterozygous CCR5 Δ32 genotype and vigorous p24-stimulated peripheral blood mononuclear cell (PBMC) proliferation. Therefore, we studied in detail viral burden and immunological responses in this individual.
Methods
PBMC and gut and lymph node biopsy samples were analysed for proviral HIV-1 DNA by real-time and nested PCRs, and nef/LTR alleles by nested PCR. HIV-specific antibodies were studied by Western blotting, and CD4+ and CD8+ T lymphocyte responses were measured by proliferation and cytokine production in vitro.
Results
PBMC samples from 1996, but not since, showed amplification of nef alleles with gross deletions. Infectious HIV-1 was never recovered. Proviral HIV-1 DNA was not detected in recent PBMC or gut or lymph node biopsy samples. C135 has a consistently weak antibody response and a substantial CD4+ T cell proliferative response to a previously described HLA-DR13-restricted epitope of HIV-1 p24 in vitro, which augmented a CD8+ T cell response to an immunodominant HLA-B57-restricted epitope of p24, while his T cells show reduced levels of CCR5.
Conclusions
Subject C135's early PCR and weak antibody results are consistent with limited infection with a poorly replicating nef/LTR-deleted strain of HIV-1. With his HLA-B57-restricted gag-specific CD8 and helper HLA-DR13-restricted CD4 T cell proliferative responses, C135 appears to have cleared his HIV-1 infection 37 years after transfusion.
Keywords: HIV-1, CD4, CCR5, Nef
Introduction
Possible ‘correlates of immune protection’ in HIV-1 infection have been widely reviewed [1–4]. However, from tens of millions of individuals living with HIV globally, there is only one documented case of apparent clearance of HIV-1, the Berlin Patient, who underwent allogeneic bone marrow transplantation from a CCR5 Δ32 homozygous donor [5,6].
Earlier, after approximately 10 years of the epidemic, we originally observed a small group of individuals living with HIV-1, the Sydney Blood Bank Cohort (SBBC), who remained asymptomatic [7]. These subjects were subsequently found to be infected with a nef/long terminal repeat (LTR)-deleted strain of HIV-1 [8], and attenuation of the transmitted viral strain appeared to be the common factor in this asymptomatic cohort. This is consistent with data from experimental infection with nef-deleted simian immunodeficiency virus (SIV) [9], but overall, nef deletion is not a common finding in cohorts of long-term non-progressors (LTNPs) [10–12]. Alternatively, LTNP cohorts exhibit over-representation of CCR5 Δ32 heterozygous individuals [13–15], implicating reduced viral co-receptor expression in limiting viraemia. In the case of the Berlin Patient, it is plausible that ablative therapy and transplantation with progenitors that lacked expression of CCR5 were the main reason for the dramatic reduction of viral burden, although it cannot be ruled out that a low-grade graft versus host immune response could also have played a role [5], since other studies found that such protocols reduce HIV DNA in peripheral blood mononuclear cells (PBMCs) to below detectable levels, but ultimately recipients’ HIV plasma viraemia can rebound in the absence of antiretroviral therapy (ART) from an unknown reservoir [16,17].
However, there is clear evidence for immune control of viral burden, as indicated by the prevalence of HLA-B27 and HLA-B57 genotypes amongst LTNP [18]. Up to half of LTNPs with undetectable plasma viral load, in the absence of ART, referred to as Elite Controllers, are HLA-B57 [19–21], consistent with the consensus that HIV-specific CD8+ cytotoxic T lymphocytes (CTLs), particularly Gag-specific CTL, contribute to control of viraemia [3]. Another select group of LTNP subjects includes those with unusually strong CD4 lymphoproliferative responses to p24 [22,23], which may also exert anti-p24 cytotoxicity [24,25]. Furthermore, one report suggested that individuals with HLA-DRB1*13 genotype have higher CD4 lymphoproliferative responses to p24 and associated control of viraemia [26], while HLA-DRB1*13 was over-represented in elite controllers [27]. HLA-DRB1*1303 and HLA-DRB1*1302 were also found to be associated with lower plasma viral loads in two other studies [28,29].
We have investigated immunological and virological parameters in one particular individual, subject C135, who was transfused with the SBBC strain of nef/LTR-deleted attenuated HIV-1, as detailed previously [30], and maintained a steady CD4 T cell count, unlike some other subjects in this cohort [31,32]. Subject C135 is also CCR5 Δ32 heterozygous [31], has HLA-B57 and HLA-DRB1*13:02:01 alleles [23], and a vigorous PBMC proliferative response to HIV-1 p24 [23,33]. Therefore, in this individual, five well-defined factors associated with LTNP are combined. We now show that C135 has a potent CD4+ T cell response to an HLA-DR13-restricted epitope in HIV-1 p24 in vitro, which in turn appears to assist CD8+ T cells to proliferate in response to HLA-B57-restricted epitopes of p24. These responses are associated with complete control over replication of HIV-1 in vivo for over 30 years, in the absence of ART, to the point where cells latently infected with HIV-1 DNA can no longer be detected in PBMC, or in biopsy samples of either lymph node or gut-associated lymphoid tissue.
Methods
Subjects
Subject C135, a 72-year-old male, is a member of the SBBC who was transfused in 1981 with a unit of erythrocytes from donor D36, later shown to be infected with nef/3′ LTR-deleted virus [30,31]. Other SBBC transfusion recipients from this donor were also LTNPs [7,8,31,32], consistent with attenuation of the transmitted viral strain. C135 was identified in 1996 by tracing of recipients of transfusions from donor D36 and remains completely asymptomatic after a total of 37 years. Detailed serological findings for this subject are described in the results section. C135 is CCR5 Δ32 heterozygous [31] and HLA A1,33; B50,57; DR7,13 [23]. The HLA-DRB1 alleles, by high-resolution typing, are 07:01:01 and 13:02:01.
Healthy adult controls were recruited from university and hospital staff. The study was approved by the Australian Red Cross Blood Service–New South Wales Institutional Ethics Committee, and written informed consent was obtained.
HIV-1 Western blots
Prospectively collected longitudinal serum samples from C135 were examined for antibodies to HIV-1 using the BioRad New LAV Blot I (BioRad, Marnes La Coquette, France), according to the manufacturer's instructions.
Amplification of HIV-1 proviral DNA
Proviral DNA was amplified from patient PBMC DNA using an ultrasensitive PCR method based on a biphasic booster amplification protocol and nef5′ and LTR3′ primers, and the nef/LTR product was cloned and sequenced, as previously described [30,34].
In addition, five other assays for HIV-1 proviral DNA have been used, including (1) the diagnostic Roche Amplicor Monitor HIV DNA PCR, with a reported sensitivity of 5 copies per 10,000 PBMCs (Roche Diagnostics, Basel, Switzerland); (2) a published PCR assay for HIV-1 gag DNA with an estimated sensitivity of 10 copies per 1,000,000 cells [35]; (3) a published sensitive real-time PCR for HIV-1 DNA using gag primers, capable of detecting <10 copies per 300,000 cells [36,37]; (4) a published real-time PCR for HIV-1 DNA using pol primers, capable of detecting <3 copies per 100,000 cells [38,39] [40]; and (5) nested PCR for HIV-1 gag [41] with a sensitivity <<10 copies per 300,000 cells [42].
DNA for assays 3–5 was extracted using the Qiagen DNeasy Blood and Tissue kit (Qiagen, Hilden, Germany).
The single-copy assay was used to quantify plasma viral load as previously described [43].
Altogether, these assays were performed in four independent laboratories.
Lymphocyte immunophenotyping and cell sorting
Lymphocyte immunophenotyping of fresh whole-blood samples from C135 between 2009 and 2014 was performed as previously described [44,45].
Memory CD45RO+ CD4+ cells for HIV DNA analysis were purified from PBMC using a FACSAria (BD Biosciences, San Jose, CA, USA), as previously described [37,39]. Similarly, CD4+ T cells were also purified from gut and lymph node biopsy samples (see further) by cell sorting.
Lymphocyte function assays
From 1996 to 2006, PBMCs from C135 were assayed for proliferation in response to HIV-1 p24 in a 6-day 3H-thymidine uptake assay, as previously described [33]. PBMCs were also labelled with either carboxyfluorescein diacetate succinimidyl ester (CFSE) or Cell Trace Violet (Thermo Fisher Scientific, Waltham, MA USA), and cultured in the presence or absence of recombinant HIV-1 p24, or with pools of overlapping peptides or individual peptides from the sequence of HIV-1 Gag, as previously described [23,46,47]. Proliferation was optimised by supplementation at day 5 of culture with low doses of exogenous interleukin (IL)-15 (0.5 ng/mL; R&D Systems, Minneapolis, MN, USA) and IL-21 (0.1 ng/mL; BioSource International, Camarillo, CA, USA), followed by a further 5 days of culture. This culture system has been extensively validated to provide an optimal ratio of antigen-specific proliferation to background proliferation [48]. At the end of the culture, PBMCs were stained with CD3-PerCP-Cy5.5 or -Pacific Blue, CD4-Alexa Fluor 700 and CD8-APC-Cy7, CD25-APC and CD71-PE (BD Biosciences, San Jose, CA, USA) and analysed on a four-laser LSR II (BD Biosciences), as previously described [48]. All proliferation experiments contained negative control cultures in the absence of added HIV-1 Gag antigenic peptides and positive control cultures containing phytohaemagglutinin (PHA), respectively, and were performed on at least three occasions.
Intracellular cytokine (ICC) assays, in response to HIV-1 Gag peptides, were performed using fresh sodium heparin-anticoagulated whole blood, as previously described [36,49], with a validated cut-off for positive results of 0.08% (mean from HIV uninfected healthy adult controls (n = 20) plus 3× SD) [50]. All ICC experiments included negative control cultures and positive control cultures containing Staphylococcal enterotoxin B (SEB), respectively, and were performed on at least two occasions.
Lymph node and gut biopsies
Ultrasound-guided fine needle biopsy (FNB) was performed in 2014 under local anaesthesia, as previously described [51], using a 25-gauge needle inserted into a single lymph node in the inguinal chain. Cells were transferred into RPMI/10% FCS, pelleted by centrifugation and resuspended, and CD4 T cells were accurately counted using immunophenotyping in TrucountTM tubes (BD Biosciences), as previously described [51]. The remaining cells were cell sorted for CD4+ T cells for HIV DNA analysis.
C135 underwent routine endoscopy and colonoscopy screening in 2009 and provided 10 pinch biopsy samples taken from each of three sites: left colon, right colon and terminal ileum. No abnormalities were detected during the procedure. Single-cell suspensions were prepared from these biopsy samples, as previously described [44,45]. CD45+ CD4+ lymphocytes and CD45–epithelial cell adhesion molecule (EpCAM)+ epithelial cells were counted using TrucountTM tubes. Remaining cells were cell sorted for CD4+ T cells for HIV DNA analysis.
CCR5 expression on memory CD4+ and CD8+ T lymphocytes
Fresh whole-blood samples from C135 and healthy adult controls, respectively, were stained exactly in parallel within 1 hour of venepuncture using an optimised indirect immunofluorescence method for CCR5, as previously described [52,53]. Briefly, 100 μl of whole blood was incubated with 1 μg of purified anti-CCR5 monoclonal antibody (mAb), clone 2D7 (BD Biosciences) for 1 hour on ice, washed twice with PBS, incubated with 1/50 dilution of PE-labelled affinity purified Fab′2 goat anti-mouse IgG (H + L chain) (Jackson ImmunoResearch, West Grove, PA), washed once, incubated with 1/10 dilution of normal mouse serum for 10 minutes at RT, then incubated for a further 15 minutes at RT with directly conjugated CD3, CD4, CD8 and CD45RO-ECD (Beckman Coulter, Hialeah, FL) mAb. Red cells were lysed with Optilyse C (Beckman Coulter), washed once with PBS, fixed with 0.5% paraformaldehyde in PBS and analysed on the LSR II flow cytometer, as described earlier.
Statistical analysis
Results are expressed as medians and interquartile ranges, analysed using Prism versions 6 and 7 (GraphPad, La Jolla, CA, USA).
Results
HIV serology and CD4 T cell counts
C135 presented in 1996 (Figure 1a), 15 years after infection, as HIV antibody positive by routine diagnostic enzyme immunoassay, but with an indeterminate Western blot (WB) pattern [30], as shown in Figure 1b, according to standard Australian interpretative guidelines [54]. An indeterminate WB serology was maintained for all available samples, with weak p18, p24 and gp160 bands between 1996 and 2007. These results would be interpreted as HIV positive, according to Centers for Disease Control and Prevention guidelines [55], and also in combination with positive nucleic acid testing for HIV-1 on at least two occasions (see further), by revised Australian criteria [56].
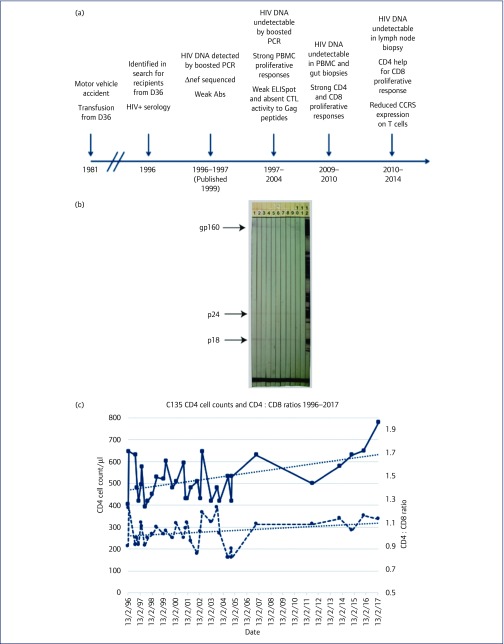
Figure 1.
Timeline of studies, HIV-1 serology and CD4 T cell counts for subject C135. (a) Timeline of transfusion, testing, sampling and studies for C135. See text for details. (b) Western blot analysis of longitudinal serum samples from C135, consistently showing faint bands at p18, p24 and gp160, for the following dates: 25/9/96 (lane 1), 14/4/97 (lane 2), 14/7/98 (lane 3), 23/8/99 (lane 4), 17/10/00 (lane 5), 23/8/01 (lane 6), 1/10/02 (lane 7), 23/9/03 (lane 8), 4/11/04 (lane 9), 29/6/07 (lane 10), kit negative control (lane 11), and kit positive control (lane 12). Note that the bands on the original blots are faint to the naked eye but easily visible. The level of brightness of the original digital photographic image has been reduced by 25% to make bands easier to visualise for this report, but makes the bands on the positive control (lane 12) appear less distinct than those on the original blots. (c) Longitudinal CD4 T cell counts (solid line) and CD4 : CD8 ratios (dashed line) for C135 from 1996 to 2017. Trend lines (linear regression) are shown for each data series. CTL: cytotoxic T lymphocyte; ELISpot: enzyme-linked absorbent immunospot; PBMC: peripheral blood mononuclear cell
Later WB results from five samples between 2011 and 2017 have been weakly positive for gp160, with all other bands negative. In contrast, the other SBBC members had full WB reactivity [7,30]. The WB pattern for C135 is uncommon and can be seen in some primary HIV subjects early during seroconversion [54].
CD4 T cell counts for C135 over the 21 years between 1996 and 2017 remained steady (median: 505 cells/μl), as did the CD4 : CD8 ratio (median: 1.03), as shown in Figure 1c.
Activated CD38+ HLA-DR+ cells were 0.6%, 1.9% and 1.2% of CD8 T cells on three occasions, respectively, between 2009 and 2014 [compared to a median of 1.5% (interquartile range: 1.0%–2.0%) for 54 healthy adult controls in a contemporaneous study [49]]. Therefore, C135's PBMCs have shown no change in CD8 activation levels since he had a median of 2.0% from nine samples between 1996 and 1998, in our earlier report in 1999 [57].
Thus, C135 differs from other LTNP subjects with nef-deleted HIV-1, whose CD4 cell counts have eventually declined [58], including all but two other members of the SBBC [31].
HIV-1 nef/LTR DNA PCR assays and sequencing
Proviral HIV-1 DNA was originally detected in PBMC from C135 in 1996, and the number of copies was estimated to be 2–5 copies per 100,000 PBMCs [30]. Longitudinal PBMC samples from C135 were screened for the presence of the nef/LTR region of proviral HIV-1 using a boosted PCR (Table 1), as previously described [30,34]. Four samples from 1996 and 1997 were positive; however, all subsequent samples tested were negative by the boosted PCR (Table 1).
Table 1.
PCR amplification of proviral DNA encoding the nef/LTR region
| Sample date | nef/LTR PCR result | Length of nef/LTR bp |
|---|---|---|
| 13.02.96 | − | na |
| 11.03.96 | − | na |
| 09.05.96 | + | 660 |
| 25.09.96 | + | 664 |
| 18.11.96 | + | 664 |
| 24.03.97 | + | 660 |
| 14.07.97 | − | na |
| 04.11.98 | − | na |
| 19.10.99 | − | na |
| 28.02.00 | − | na |
| 23.05.01 | − | na |
| 08.05.02 | − | na |
| 05.06.04 | − | na |
na: not applicable.
Amplification of samples from 1996 to 1997 with primers nef5′ and LTR3′ gave 660, 664-, 664- and 660-bp products, respectively, on the occasions for which a positive signal was obtained (Table 1), compared with an 882-bp product for HIV-1NL4-3. Cloning and sequencing of the nef/LTR amplimer (25.09.96) showed deletions of 1, 72, 4 and 11 bp from the nef-alone region relative to HIV-1NL4-3, and 139 bp from the nef/LTR overlap. This 139-bp deletion includes the deletion common to all SBBC strains [30,34], equivalent to nucleotides 9281–9435 of HIV-1NL4-3, and includes the 5′ nuclear factor (NF)-κB binding sequence. Other features common to the SBBC strain of viruses include a GA-rich sequence block at a position equivalent to HIV-1NL4-3 9258–9270 bp, together with duplicated NF-κB and Sp1 sequences, which differ in arrangement between cohort viruses [8,30,34]. The sequence of C135 includes an additional Sp1 and a partial NF-κB binding sequence [30].
The C135 sequence encodes a truncated Nef open reading frame of 19 amino acids due to the introduction of a termination codon (TAA) brought into phase by the deletion of a single nucleotide equivalent to nucleotide 8842 of HIV-1NL4-3 [30]. However, translation beginning at a second ATG codon (at nucleotides 8822–8824 with respect to HIV-1NL4-3), in a different reading frame, would encode a protein containing 96 amino acids of Nef, although, of the conserved blocks, only a complete block B would be retained.
HIV DNA PCR assays of peripheral blood mononuclear cells and purified CD4 T cells
Table 2 summarises PCR results from C135 from 2002 onwards, from samples including PBMC, whole blood, and purified CD4 T cells from PBMC, gut and lymph node biopsies.
Table 2.
PCR results for C135 samples
| Sample | Date | PCR | Amount of sample | Result | Limit of detection | Method reference |
|---|---|---|---|---|---|---|
| PBMC | 2002–2004 | gag DNA PCR | 5 × 106 cells | Negative | <10 copies/106 cells | [35] |
| Whole blood | 2006–2007 | Roche Qualitative Monitor v1.5 | 0.5 mL whole blood | Negative | 5 copies/10,000 cells | Roche product insert |
| Gut biopsy CD4 T cells | August 2009 | gag DNA PCR | 156,000 cells | Negative | <10 copies/300,000 cells | [36,37] |
| PBMC | March 2010 | gag DNA PCR | 83,000 | Negative | <10 copies/300,000 cells | [36,37] |
| Memory CD4 T cells from PBMC | March 2010 | gag DNA PCR | 2 × 106 cells | Negative | <10 copies/300,000 cells | [36,37] |
| PBMC | September 2014 | pol DNA PCR | 6.23 × 106 cells | Negative | <3 copies/100,000 cells | [40] |
| Lymph node CD4 T cells | September 2014 | nested gag PCR | 76,000 cells | Negative | <<10 copies/300,000 cells | [42] |
| Plasma | April 2014 | gag RNA PCR | 14 ml | Negative | <0.3 copy/mL | [43] |
PBMC: peripheral blood mononuclear cell.
HIV DNA PCR testing of fresh whole blood, using samples from 2006 and 2007, were negative by the qualitative diagnostic Roche Amplicor HIV-1 DNA test.
Negative results were also obtained for fresh PBMC samples by three other assays. The first PCR assay for HIV-1 gag DNA in PBMC had an estimated sensitivity of 10 copies per 1,000,000 cells [35]. This assay readily detected HIV DNA in PBMC from other members of the SBBC (NK Saksena, unpublished data).
The second assay, a real-time PCR for HIV-1 DNA in PBMC using Gag primers, capable of detecting <10 copies per 300,000 cells [36,37], was also negative. This assay was also negative on 2 × 106 highly purified memory CD45RO+ CD4 T cells, but this assay readily detected HIV DNA in PBMC and cell-sorted CD4 T cells from other HIV-positive subjects [36,37].
The third assay was a real-time PCR for HIV-1 DNA using pol primers, capable of detecting <3 copies per 100,000 cells [38,39]. A total of 53 replicates were assayed using this assay, from an equivalent of 6.23 × 106 PBMCs, and all replicates were negative.
Furthermore, plasma samples have always remained negative by routine diagnostic HIV-1 plasma viral load assays [31,33] and were also negative in 2014 using the single-copy assay [43]. The latest diagnostic HIV viral load result from 2017 was <20 copies/mL.
Importantly, HIV-1 has never been isolated from PBMC samples from C135, despite numerous attempts [30,59], using coculture techniques that resulted in successful isolation of viral strains from other members of the SBBC [7,8,59].
Lymph node and gut biopsies
Lymph node cells were obtained from C135 by ultrasound-guided FNB of an inguinal lymph node in 2014. The number of CD4 T cells recovered was 148,043 cells, which was towards the low end of the observed normal range for inguinal lymph node FNBs from 10 healthy adult controls (73,701–1,666,592), as previously described [51].
HIV DNA in cell-sorted FNB CD4 T cells was negative using a nested gag PCR [41], recently optimised for assays of antigen-specific CD4 T cells [42].
CD4+ T lymphocytes were also isolated from gut biopsy samples in 2009. The number of CD4 T cells in pooled single-cell suspensions derived from biopsy samples from terminal ileum, left colon and right colon for C135 was 28,415 CD4 T lymphocytes/106 EpCAM+ epithelial cells, similar to the median for results for these three sites, from 21 HIV-uninfected adults of 26,381 CD4 T lymphocytes/106 EpCAM+ epithelial cells, as previously described [45].
HIV DNA was negative in the 156,000 cell-sorted C135 gut biopsy CD4 T cells (Table 2) using real-time PCR [37].
Lymphocyte function assays
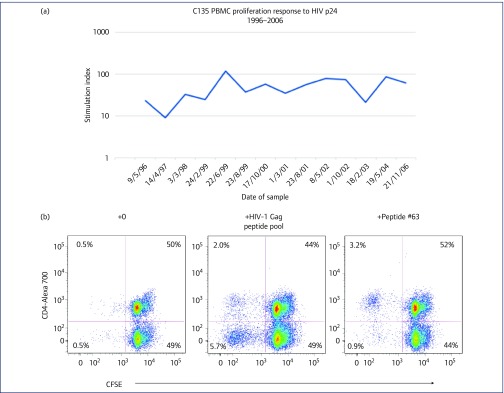
PBMC from C135 consistently showed a strong p24-specific proliferative response from 1996 to 2006 (Figure 2a), as previously described [23,33,60]. We now confirm that this response was due to proliferation of CD4+ T lymphocytes (Figure 2b), which is characteristic of LTNP [22,61]. Furthermore, when responses to individual HIV-1 Gag peptides were studied previously [23], the PBMC proliferative response was found to be directed to the 15-mer WMTNNPPIPVGEIYK, which we now also confirm as a CD4 proliferative response (Figure 2b). This sequence contains an epitope previously reported to be presented on HLA-DRB1*1301 [26], and C135, by high-resolution typing, has the HLA-DRB1 alleles 07:01:01 and 13:02:01.
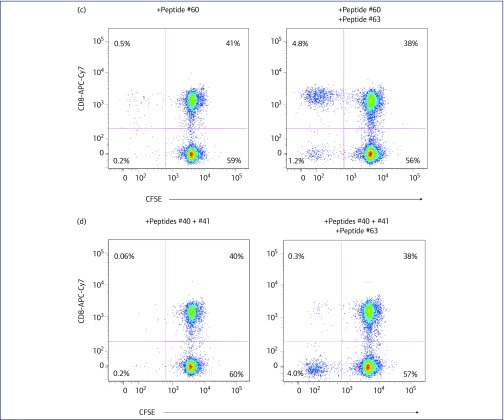
Figure 2.
CD4 and CD8 T cell responses to HIV-1 Gag antigens for subject C135. (a) Stimulation indices for C135's PBMC proliferation in response to HIV p24, for longitudinal samples from 1996 to 2006, using the 3H-thymidine uptake assay. (b) Flow cytometric analysis showing proliferating CD4+ T lymphocytes in CFSE-labelled PBMC from C135, in response to culture with HIV-1 Gag peptide pool (middle histogram) or with peptide #63, WMTNNPPIPVGEIYK (right histogram), compared with culture in the absence of antigen (left histogram). (c) Flow cytometric analysis showing proliferation of CD8+ T lymphocyte in CFSE-labelled PBMC from C135, in response to culture with the CD8 Gag peptide #60 (AGTTSTLQEQIGWMT) in the presence (right histogram) and absence (left histogram) of the CD4 peptide #63 (WMTNNPPIPVGEIYK). (d) Flow cytometric analysis showing proliferation of CD8+ T lymphocyte in CFSE-labelled PBMC from C135, in response to culture with the CD8 Gag peptides #40 (KVVEEKAFSPEVIPM) and #41 (EKAFSPEVIPMFSAL) in the presence (right histogram) and absence (left histogram) of the CD4 peptide #63 (WMTNNPPIPVGEIYK). CFSE: carboxyfluorescein diacetate succinimidyl ester; PBMC: peripheral blood mononuclear cell
Moreover, PBMC also showed a CD8 T-lymphocyte proliferative response to the pool of 123 HIV-1 Gag overlapping peptides (see Figure 2b, middle histogram, bottom left quadrant). Such a CD8 proliferative response is characteristic of HLA-B57 LTNP [62]. We attempted to map CD8+ T-lymphocyte responses to individual peptides, but only minimal CD8+ responses were found in the absence of a concurrent CD4 response (see left histograms, Figures 2c,d), even culturing optimally with the addition of low doses of exogenous IL-15 and IL-21 [48]. However, a clear CD8 proliferative response was observed when PBMC were incubated with the 15-mer peptide containing the well-characterised HLA-B57 p24 epitope, TSTLQEQIGW [63,64], in the presence of the CD4 response to the CD4-specific p24 peptide, WMTNNPPIPVGEIYK (Figure 2c). Another well-described HLA-B57 epitope, KAFSPEVIPMF [63], also gave a very weak CD8 proliferative response, again only in the presence of the CD4 antigenic peptide WMTNNPPIPVGEIYK (Figure 2d).
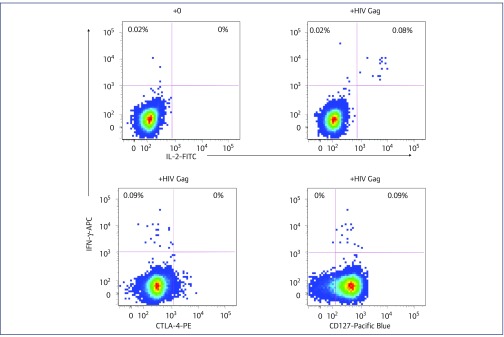
Previous experiments with PBMC from C135 have shown weak interferon (IFN)-γ responses and absent CTL responses immediately ex vivo [23,33,65]. Using the ICC assay, we found a small, but distinct, CD4 IFN-γ+ response to HIV-1 Gag peptides (Figure 3), which represented around 0.1% of CD4+ T cells in fresh whole blood. Even though this is a relatively low level of antigen-specific CD4+ T cells, it still reaches our validated cut-off for positive ICC results (0.08%), determined for HIV-uninfected controls as part of a vaccine trial [50]. The population of IFN-γ+ CD4+ T cells clearly also made IL-2 and were CD127 positive (IL-7Rα+), but were CTLA-4 negative (Figure 3), consistent with their proliferative phenotype.
Figure 3.
Intracellular cytokine assays. Antigen-specific CD4+ T cells by intracellular cytokine assay for subject C135. Intracellular flow cytometric analysis of the CD4+ T cell cytokine response in fresh whole blood from C135, cultured with a pool of overlapping HIV-1 Gag peptides, showing the production of IFN-γ and IL-2, the lack of production of the inhibitory receptor CTLA-4, in combination with the expression of the IL-7Rα chain, CD127, a marker of long-term memory T cells. Background production of IFN-γ and IL-2 in the absence of HIV-1 Gag peptides is shown in the upper left histogram. IFN: interferon; IL: interleukin
Cell surface CCR5 expression
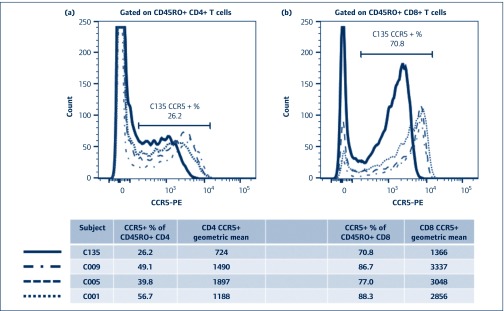
C135 is CCR5 Δ32 heterozygous [31], and we examined expression of CCR5 on T cells in fresh whole blood, as shown compared with T cells from healthy adult controls, stained and analysed in parallel (Figure 4). The results show that C135 has a demonstrably lower level of CCR5 expression on his CD4+ T cells (Figure 4a), but not a dramatically lower percentage of CCR5+ CD4+ T cells. The difference is even clearer when CD8+ T cells are examined (Figure 4b), again with a lower expression level seen as a lower fluorescence intensity, but not as a lower CCR5+ percentage.
Figure 4.
Relatively low level of CCR5 expression on CD4 and CD8 T lymphocytes from subject C135. (a) Flow cytometric histogram of CCR5 expression on CD4+ T lymphocytes from C135 (solid black line) and three healthy adult controls (dashed histograms), respectively. (b) Flow cytometric histogram of CCR5 expression on CD8+ T lymphocytes from C135 (solid black line) and three healthy adult controls (dashed histograms), respectively
It should be noted that PBMC from C135 were able to be infected with laboratory strains of HIV-1 in vitro, although this required depletion of inhibitory CD8+ lymphocytes [23].
Discussion
Subject C135 is distinct from other members within the highly unusual SBBC, infected with the same nef/LTR-deleted HIV-1, and compared with other elite controllers. For over 37 years, C135 has maintained undetectable plasma viral load, an uncommon indeterminate WB pattern, and steady CD4 cell count and CD4 : CD8 ratio within the normal range, without CD8 activation.
What really sets C135 apart as unique is having had detectable HIV DNA in PBMC samples from around 15 years after infection, but subsequent negative results on a range of assays of HIV DNA in PBMC, purified memory CD4 T cells and lymph node and gut biopsy samples. In contrast, two of the other SBBC recipients, C49 and C64, with consistently undetectable plasma viral loads without ART, have a readily detectable reservoir of HIV-1 DNA in PBMC, and C64 has had virus isolated on one occasion from PBMC [31,34,59] (and NK Saksena, unpublished data). A leukapheresis sample has not been available from C135 but could possibly be tested in future, more exhaustive studies of viral outgrowth assays of replication-competent HIV-1.
One study of PBMC from patients on ART reported that two out of 30 were negative for HIV DNA, but all patients had HIV DNA detected in rectal CD4 cells [66]. Three early studies of elite controllers reported only one negative HIV DNA PCR result out of a total 25 elite controllers [11,20,21], but analysis of tissue samples was not reported. One large study of 46 controllers found only one subject negative using a sensitive assay for HIV RNA in plasma, and this subject was positive for proviral DNA in PBMC [67]. Another study of four highly selected elite controllers, with weak indeterminate HIV WBs similar to C135, has also found very low quantitative levels of viral burden, but with at least one positive assay of HIV DNA from PBMC, or from biopsy samples of colon or ileum, as well as one instance of viral outgrowth in high input coculture assay [68]. In these latter four cases, proliferative CD8 responses to HIV-infected autologous CD4 T cells and granzyme B-associated cytotoxicity were a feature, but CD4 proliferative responses were not reported. Only two of these cases were HLA-B57 and one had some evidence of a nef deletion but still had replication-competent viral outgrowth [68]. Furthermore, another study of elite controllers reported significantly elevated activation of circulating CD8 T cells [69]. Altogether, subject C135 differs from other elite controllers in the literature.
The combination of the antibody results and the T-lymphocyte HIV Gag peptide-specific proliferation and cytokine responses, which are consistent with this subject's HLA typing, and together with the early PCR results, convincingly argues that C135 has been weakly infected with a poorly replicating HIV-1 strain. C135 has no other risk factors, and it is believed that he has not been exposed to HIV-1 except in the case of the transfusion in 1981 with an HIV-infected unit from the SBBC donor D36 [31]. Furthermore, consistent longitudinal results argue strongly against any mistaken identity or laboratory error in a single sample. Additionally, the sequencing data from the early PCR positive samples from C135 yielded a sequence distinct from, but still closely related to, sequences obtained from other SBBC cohort samples, further arguing against simple contamination or mistaken samples.
We and others have found that primary HIV-1 infection in subjects who undergo treatment initiation very early can demonstrate a weakening of their HIV-specific antibody levels [70–73]. More recently, it has been reported that HIV-positive patients who underwent allogeneic stem cell transplantation and subsequently exhibited undetectable HIV DNA showed declining titres of HIV-1 antibodies [17,74]. Similarly, the Berlin Patient has exhibited a decline in HIV-specific titres [6], and while it was interpreted that such antibody titre declines may have been due to lack of antigen, in these allogeneic stem cell transplant cases, this was possibly confounded by the conditioning regimens. Altogether, in these cases, and in elite controllers [67,68], low antibody levels correlated with unusually low levels of viral burden.
The limited development of the anti-HIV-1 antibody response in C135 and the very low HIV DNA proviral load suggest that HIV-1 replication was restricted very early after infection, consistent with previous observations of low viral load in HLA-B57 individuals during acute infection [64]. Also, the limited antibody response to HIV envelope proteins suggests that neutralising antibodies played a minor role, if any, in limiting infection. In fact, plasma samples from C135 showed little or no activity when tested for neutralisation of a range of clade B isolates [59].
C135 has a CD4 cell count and a CD4 : CD8 ratio in the normal range, and within those CD4+ T cells, along with other SBBC members, has a high proportion of circulating memory phenotype cells [57]. We tested purified memory CD4 T cells from C135's PBMC and obtained a negative PCR result. We also tested CD4+ T cells from lymphoid tissue and gut biopsy samples from C135, which were negative. Using the same methods, for samples from other subjects living with HIV, we have previously readily measured HIV DNA in memory CD4 T cells from PBMCs [36,37,39,41], gut biopsy samples [44,45] and lymph node fine needle biopsy samples [51].
Another reservoir that may harbour infected cells over such a long period could be central nervous system (CNS), but sampling of this compartment is problematic, and we have evidence that even mild symptomatic CNS involvement is reflected in HIV DNA levels in circulating CD4 T cells [40]. The transfusion donor, D36, developed HIV-associated dementia after 18 years of being infected with the same strain of nef/LTR-deleted HIV-1 [34]. However, donor D36 had a detectable plasma viral load and declining CD4 cell count [31] and elevated CD8 T cell activation, in contrast with C135 [57]. The undetectable viraemia, steady CD4 cell count and lack of activation of CD8 T cells, over more than three decades, argues against a smouldering infection in a non-lymphoid reservoir in C135.
C135 has a CCR5 Δ32 heterozygous genotype and reduced cell surface expression of CCR5, which is known to be associated with LTNP status [13,15]. Furthermore, the nef/LTR-deleted strain of HIV-1 with which he was transfused has clearly been shown to be attenuated in vivo [8]. Nef is believed to reduce cell surface expression of HLA class I molecules, and its lack may have helped the Gag-specific CD8 CTL in C135 to rapidly identify infected cells and limit viral burden early in infection [64,75]. One report has described that, amongst rhesus macaques immunised with nef-deleted SIV, those macaques with major histocompatibility complex class I alleles previously known to be associated with control of SIV infection exhibited better subsequent control of acute infection with heterologous strains of SIV [76], suggesting synergy between nef deletion and CD8 responses, analogous to subject C135.
The present studies of PBMC proliferation demonstrate that CD4 T cells from C135 may provide help for potent immunological HLA-B57-restricted CD8 T cell control of HIV-1 infection. C135 had responses to two of the best-studied HLA-B57 CD8+ CTL epitopes in p24. The proliferative CD4 response to the sequence WMTNNPPIPVGEIYK adjacent to helix 7 has previously been reported to be associated with viral control in HLA-DRB1*1301 individuals [26]. While much effort has been made to delineate protective CD8 T cell responses, proliferative Gag-specific CD4 T cell responses are even more highly associated with LTNP status [22,61]. Previously we found that PBMCs from C135 have shown weak IFN-γ responses and absent CTL responses immediately ex vivo [23,33,65]. In contrast with the IFN-γ and CTL results, we had previously found that PBMCs from C135 exhibited strong in vitro CD8 antiviral suppressive activity [23], which has also been reported for other HIV-positive LTNP and elite controllers [23,77–79]. Furthermore, PBMCs from C135 protected reconstituted severe combined immunodeficiency mice from superinfection with HIV-1, in an adoptive transfer model [80].
C135's CD4 response augmented his CD8 response to HLA-B57-restricted HIV-1 Gag peptides. The first CD8 epitope, TSTLQEQIGW, has been described to mutate relatively readily, but with a fitness cost [81]. The other small C135 CD8 response is to the epitope, KAFSPEVIPMF, which is located in the highly conserved helix 2 of the N-terminal domain of p24, and mutation of a proline in this helix rendered HIV-1 non-infectious [82]. We, and others, have described LTNP subjects with very large populations of proliferative CD4+ CTL that also targeted a nearly identical epitope in helix 2 [24,83]. These results are similar to a previous study where IL-2 produced by CD4 T cells stimulated with an HIV-1 Nef peptide dramatically enhanced a CD8 response to a different Nef peptide [84], but the CD4 enhancing activity was lost after resolution of acute infection. For subject C135, the CD4 proliferative activity has been maintained for over 30 years, as also reported for memory CD4 T cell responses to vaccinia virus [85].
While it would be desirable to design vaccines to stimulate appropriate targeting of conserved residues in helices 2, 6 and 7 of HIV p24 [86], it is difficult to conceive how non-HLA-B57, -B27 or -DR13 subjects could be vaccinated against exactly the same epitopes. Yet helices 2, 6 and 7 and the cyclophilin binding loop may represent a potential therapeutic target for a small molecule [87]. However, a therapeutic approach to block Nef function is completely unclear at present, even though nef deletion appears to be the best described attenuation for either HIV-1 or SIV in vivo, and inhibition of Nef may be critical to T cell control of the HIV-1 reservoir [88].
We interpret the results for C135 as a probable case of clearance of HIV-1 infection, resulting from a combination of plausible immune, host genetic and viral attenuation mechanisms. However, with only a single individual subject, caution is still required yet provides some hope that a similar combination of favourable pressures can be more generally applied to HIV-1 infection with a similar outcome.
Acknowledgements
The authors thank the subject C135 for his co-operation in providing samples, Julie Yeung and Kate Merlin for PBMC cryopreservation, Leon McNally and Alex Carrera for diagnostic PCR assays, as well as the NIH Reference Reagents Program for provision of HIV-1 overlapping Gag peptides.
This project was partly funded by Australian National Health and Medical Research Council Fellowships (1063422 to JZ, 351041 to CMLM), Project Grants (101085 to JZ) and Program Grants (510448 and 1052979 to AK, DAC). WBD was supported by The Australian Red Cross Blood Service, an NHMRC project grant, and the Australian Centre for HIV/HCV Virology Research. SP and BH were supported by the Delaney AIDS Research Enterprise (DARE) to Find a Cure (1U19AI096109 and 1UM1AI126611-01), Australian Centre for HIV and Hepatitis Virology Research (ACH2), and the Australian National Health and Medical Research Council (Project Grant AAP1061681).
Authors’ contributions
JZ, WBD, MC, CMLM, PHC, KS, BW, SP, PRG, NS, NKS and AK designed experiments, and JZ, WBD, MC, CMLM, PC, KS, BW, KMcB, WH-N, BH, MB and YX performed experiments. MD recruited and performed endoscopy. SR provided clinical care for subject C135 and identified the clinical possibility of viral clearance. AK, KK, DAC, JSS and JL supervised the recruitment and management of LTNP cohorts. JZ, WBD, MC, CMLM, PHC, KK, SP and AK analysed the data. JZ, WBD, MC, CMLM and AK vouch for the data and analysis and wrote the paper.
Conflict of interest
The authors do not disclose any financial conflicts of interest.
References
- 1. Pantaleo G, Koup RA. Correlates of immune protection in HIV-1 infection: what we know, what we don't know, what we should know. Nat Med 2004; 10: 806– 810. [DOI] [PubMed] [Google Scholar]
- 2. Heeney J, Plotkin SA. Immunological correlates of protection from HIV infection and disease. Nat Immunol 2006; 7: 1281– 1284. [DOI] [PubMed] [Google Scholar]
- 3. Walker BD, Yu XG. Unravelling the mechanisms of durable control of HIV-1. Nat Rev Immunol 2013; 13: 487– 498. [DOI] [PubMed] [Google Scholar]
- 4. Zaunders J, Bockel D. Innate and adaptive immunity in long-term non-progression in HIV disease. Front Immunol 2013; 4: 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Hutter G, Nowak D, Mossner M et al. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N Engl J Med 2009; 360: 692– 698. [DOI] [PubMed] [Google Scholar]
- 6. Yukl SA, Boritz E, Busch M et al. Challenges in detecting HIV persistence during potentially curative interventions: a study of the Berlin patient. PLoS Pathog 2013; 9: e1003347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Learmont J, Tindall B, Evans L et al. Long-term symptomless HIV-1 infection in recipients of blood products from a single donor. Lancet 1992; 340: 863– 867. [DOI] [PubMed] [Google Scholar]
- 8. Deacon NJ, Tsykin A, Solomon A et al. Genomic structure of an attenuated quasi species of HIV-1 from a blood transfusion donor and recipients. Science 1995; 270: 988– 991. [DOI] [PubMed] [Google Scholar]
- 9. Kestler HW, Ringler DJ, Mori K et al. Importance of the nef gene for maintenance of high virus loads and for the development of AIDS. Cell 1991; 65: 651– 652. [DOI] [PubMed] [Google Scholar]
- 10. Rhodes DI Ashton L Solomon A et al. for the Australian Long-Term Nonprogressor Study Group . Characterization of three nef-defective human immunodeficiency virus type 1 strains associated with long-term nonprogression. J Virol 2000; 74: 10581– 10588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bailey JR, Williams TM, Siliciano RF, Blankson JN. Maintenance of viral suppression in HIV-1-infected HLA-B*57+ elite suppressors despite CTL escape mutations. J Exp Med 2006; 203: 1357– 1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wang B. Viral factors in non-progression. Front Immunol 2013; 4: 355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Stewart GJ Ashton LJ Biti RA et al. for the Australian Long-Term Nonprogressor Study Group . Increased frequency of CCR-5 delta 32 heterozygotes among long-term non-progressors with HIV-1 infection. AIDS 1997; 11: 1833– 1838. [DOI] [PubMed] [Google Scholar]
- 14. Magierowska M, Theodorou I, Debre P et al. Combined genotypes of CCR5, CCR2, SDF1, and HLA genes can predict the long-term nonprogressor status in human immunodeficiency virus-1-infected individuals. Blood 1999; 93: 936– 941. [PubMed] [Google Scholar]
- 15. Ioannidis JP, Rosenberg PS, Goedert JJ et al. Effects of CCR5-Delta32, CCR2-64I, and SDF-1 3'A alleles on HIV-1 disease progression: an international meta-analysis of individual-patient data. Ann Intern Med 2001; 135: 782– 795. [DOI] [PubMed] [Google Scholar]
- 16. Henrich TJ, Hanhauser E, Marty FM et al. Antiretroviral-free HIV-1 remission and viral rebound after allogeneic stem cell transplantation: report of 2 cases. Ann Intern Med 2014; 161: 319– 327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Koelsch KK, Rasmussen TA, Hey-Nguyen WJ et al. Impact of allogeneic hematopoietic stem cell transplantation on the HIV reservoir and immune response in 3 HIV-infected individuals. J Acquir Immune Defic Syndr 2017; 75: 328– 337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Carrington M, O'Brien SJ. The influence of HLA genotype on AIDS. Annu Rev Med 2003; 54: 535– 551. [DOI] [PubMed] [Google Scholar]
- 19. Migueles SA, Sabbaghian MS, Shupert WL et al. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc Natl Acad Sci U S A 2000; 97: 2709– 2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lambotte O, Boufassa F, Madec Y et al. HIV controllers: a homogeneous group of HIV-1-infected patients with spontaneous control of viral replication. Clin Infect Dis 2005; 41: 1053– 1056. [DOI] [PubMed] [Google Scholar]
- 21. Kloosterboer N, Groeneveld PH, Jansen CA et al. Natural controlled HIV infection: preserved HIV-specific immunity despite undetectable replication competent virus. Virology 2005; 339: 70– 80. [DOI] [PubMed] [Google Scholar]
- 22. Rosenberg ES, Billingsley JM, Caliendo AM et al. Vigorous HIV-1-specific CD4+ T cell responses associated with control of viremia. Science 1997; 278: 1447– 1450. [DOI] [PubMed] [Google Scholar]
- 23. Wang B, Dyer WB, Zaunders JJ et al. Comprehensive analyses of a unique HIV-1-infected nonprogressor reveal a complex association of immunobiological mechanisms in the context of replication-incompetent infection. Virology 2002; 304: 246– 264. [DOI] [PubMed] [Google Scholar]
- 24. Zaunders JJ, Dyer WB, Wang B et al. Identification of circulating antigen-specific CD4+ T lymphocytes with a CCR5+, cytotoxic phenotype in an HIV-1 long-term nonprogressor and in CMV infection. Blood 2004; 103: 2238– 2247. [DOI] [PubMed] [Google Scholar]
- 25. Norris PJ, Moffett HF, Yang OO et al. Beyond help: direct effector functions of human immunodeficiency virus type 1-specific CD4(+) T cells. J Virol 2004; 78: 8844– 8851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Malhotra U, Holte S, Dutta S et al. Role for HLA class II molecules in HIV-1 suppression and cellular immunity following antiretroviral treatment. J Clin Invest 2001; 107: 505– 517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ferre AL, Hunt PW, McConnell DH et al. HIV controllers with HLA-DRB1*13 and HLA-DQB1*06 alleles have strong, polyfunctional mucosal CD4+ T-cell responses. J Virol 2010; 84: 11020– 11029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Julg B, Moodley ES, Qi Y et al. Possession of HLA class II DRB1*1303 associates with reduced viral loads in chronic HIV-1 clade C and B infection. J Infect Dis 2011; 203: 803– 809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ranasinghe S, Flanders M, Cutler S et al. HIV-specific CD4 T cell responses to different viral proteins have discordant associations with viral load and clinical outcome. J Virol 2012; 86: 277– 283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rhodes D, Solomon A, Bolton W et al. Identification of a new recipient in the Sydney Blood Bank Cohort: a long-term HIV type 1-infected seroindeterminate individual. AIDS Res Hum Retroviruses 1999; 15: 1433– 1439. [DOI] [PubMed] [Google Scholar]
- 31. Learmont JC, Gezcy AF, Mills J et al. Immunologic and virologic status after 14 to 18 years of infection with an attenuated strain of HIV-1. N Engl J Med 1999; 340: 1715– 1722. [DOI] [PubMed] [Google Scholar]
- 32. Birch MR, Learmont JC, Dyer WB et al. An examination of signs of disease progression in survivors of the Sydney Blood Bank Cohort (SBBC). J Clin Virol 2001; 22: 263– 270. [DOI] [PubMed] [Google Scholar]
- 33. Dyer WB, Zaunders JJ, Yuan FF et al. Mechanisms of HIV non-progression; robust and sustained CD4+ T-cell proliferative responses to p24 antigen correlate with control of viraemia and lack of disease progression after long-term transfusion-acquired HIV-1 infection. Retrovirology 2008; 5: 112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Churchill MJ, Rhodes DI, Learmont JC et al. Longitudinal analysis of human immunodeficiency virus type 1 nef/long terminal repeat sequences in a cohort of long-term survivors infected from a single source. J Virol 2006; 80: 1047– 1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wang B, Mikhail M, Dyer WB et al. First demonstration of a lack of viral sequence evolution in a non-progressor, defining replication-incompetent HIV-1 infection. Virology 2003; 312: 135– 150. [DOI] [PubMed] [Google Scholar]
- 36. Zaunders JJ, Ip S, Munier ML et al. Infection of CD127+ (interleukin-7 receptor+) CD4+ cells and overexpression of CTLA-4 are linked to loss of antigen-specific CD4 T cells during primary human immunodeficiency virus type 1 infection. J Virol 2006; 80: 10162– 10172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. McBride K, Xu Y, Bailey M et al. The majority of HIV-1 DNA in circulating CD4+ T lymphocytes is present in non-gut homing resting memory CD4+ T cells. AIDS Res Hum Retroviruses 2013; 29: 1330– 1339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Althaus CF, Gianella S, Rieder P et al. Rational design of HIV-1 fluorescent hydrolysis probes considering phylogenetic variation and probe performance. J Virol Methods 2010; 165: 151– 160. [DOI] [PubMed] [Google Scholar]
- 39. Murray JM, Zaunders JJ, McBride KL et al. HIV DNA subspecies persist in both activated and resting memory CD4+ T cells during ART. J Virol 2014; 88: 3516– 3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Cysique LA, Hey-Cunningham WJ, Dermody N et al. Peripheral blood mononuclear cells HIV DNA levels impact intermittently on neurocognition. PLoS One 2015; 10: e0120488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kelleher AD, Long C, Holmes EC et al. Clustered mutations in HIV-1 gag are consistently required for escape from HLA-B27-restricted cytotoxic T lymphocyte responses. J Exp Med 2001; 193: 375– 386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hey-Nguyen W, Bailey M, Xu Y et al. HIV-1 DNA is maintained in antigen-specific CD4+ T cell subsets in patients on long-term antiretroviral therapy regardless of recurrent antigen exposure. AIDS Res Hum Retroviruses 2019; 35: 112– 120. [DOI] [PubMed] [Google Scholar]
- 43. Palmer S, Maldarelli F, Wiegand A et al. Low-level viremia persists for at least 7 years in patients on suppressive antiretroviral therapy. Proc Natl Acad Sci U S A 2008; 105: 3879– 3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Koelsch KK, Boesecke C, McBride K et al. Impact of treatment with raltegravir during primary or chronic HIV infection on RNA decay characteristics and the HIV viral reservoir. AIDS 2011; 25: 2069– 2078. [DOI] [PubMed] [Google Scholar]
- 45. Zaunders J, Danta M, Bailey M et al. CD4+ T follicular helper and IgA+ B cell numbers in gut biopsies from HIV-infected subjects on antiretroviral therapy are similar to HIV-uninfected individuals. Front Immunol 2016; 7: 438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zaunders JJ, Moutouh-de Parseval L, Kitada S et al. Polyclonal proliferation and apoptosis of CCR5+ T lymphocytes during primary human immunodeficiency virus type 1 infection: regulation by interleukin (IL)-2, IL-15, and Bcl-2. J Infect Dis 2003; 187: 1735– 1747. [DOI] [PubMed] [Google Scholar]
- 47. Seddiki N, Cook L, Hsu DC et al. Human antigen-specific CD4(+) CD25(+) CD134(+) CD39(+) T cells are enriched for regulatory T cells and comprise a substantial proportion of recall responses. Eur J Immunol 2014; 44: 1644– 1661. [DOI] [PubMed] [Google Scholar]
- 48. Munier CM, Zaunders JJ, Ip S et al. A culture amplified multi-parametric intracellular cytokine assay (CAMP-ICC) for enhanced detection of antigen specific T-cell responses. J Immunol Methods 2009; 345: 1– 16. [DOI] [PubMed] [Google Scholar]
- 49. Zaunders JJ, Munier ML, Kaufmann DE et al. Early proliferation of CCR5+ CD38+++ antigen-specific CD4+ Th1 effector cells during primary HIV-1 infection. Blood 2005; 106: 1660– 1667. [DOI] [PubMed] [Google Scholar]
- 50. Kelleher AD, Puls RL, Bebbington M et al. A randomized, placebo-controlled phase I trial of DNA prime, recombinant fowlpox virus boost prophylactic vaccine for HIV-1. AIDS 2006; 20: 294– 297. [DOI] [PubMed] [Google Scholar]
- 51. Hey-Nguyen WJ, Xu Y, Pearson CF et al. Quantification of residual germinal center activity and HIV-1 DNA and RNA levels using fine needle biopsies of lymph nodes during antiretroviral therapy. AIDS Res Hum Retroviruses 2017; 33: 648– 657. [DOI] [PubMed] [Google Scholar]
- 52. Xu Y, Weatherall C, Bailey M et al. Simian immunodeficiency virus infects follicular helper CD4 T cells in lymphoid tissues during pathogenic infection of pigtail macaques. J Virol 2013; 87: 3760– 3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Xu Y, Phetsouphanh C, Suzuki K et al. HIV-1 and SIV predominantly use CCR5 expressed on a precursor population to establish infection in T follicular helper cells. Front Immunol 2017; 8: 376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Healey DS, Maskill WJ, Howard TS et al. HIV-1 western blot: development and assessment of testing to resolve indeterminate reactivity. AIDS 1992; 6: 629– 633. [PubMed] [Google Scholar]
- 55. Anonymous. Interpretation and use of the western blot assay for serodiagnosis of human immunodeficiency virus type 1 infections. MMWR 1989; 38: 1– 7. [PubMed] [Google Scholar]
- 56. McDonald A. Revised case definition for HIV infection. Aust HIV Surveill Rep 2002; 18: 9– 10. [Google Scholar]
- 57. Zaunders JJ, Geczy AF, Dyer WB et al. Effect of long-term infection with nef-defective attenuated HIV type 1 on CD4+ and CD8+ T lymphocytes: increased CD45RO+CD4+ T lymphocytes and limited activation of CD8+ T lymphocytes. AIDS Res Hum Retroviruses 1999; 15: 1519– 1527. [DOI] [PubMed] [Google Scholar]
- 58. Greenough TC, Sullivan JL, Desrosiers RC. Declining CD4 T cell counts in a person infected with nef-deleted HIV-1. N Engl J Med 1999; 340: 236– 237. [DOI] [PubMed] [Google Scholar]
- 59. Verity EE, Zotos D, Wilson K et al. Viral phenotypes and antibody responses in long-term survivors infected with attenuated human immunodeficiency virus type 1 containing deletions in the nef and long terminal repeat regions. J Virol 2007; 81: 9268– 9278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Dyer WB, Geczy AF, Kent SJ et al. Lymphoproliferative immune function in the Sydney Blood Bank Cohort, infected with natural nef/long terminal repeat mutants, and in other long-term survivors of transfusion-acquired HIV-1 infection. AIDS 1997; 11: 1565– 1574. [DOI] [PubMed] [Google Scholar]
- 61. Zaunders J, Bockel D. Innate and adaptive immunity in long-term non-progression in HIV disease. Front Immunol 2013; 4: 95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Migueles SA, Laborico AC, Shupert WL et al. HIV-specific CD8+ T cell proliferation is coupled to perforin expression and is maintained in nonprogressors. Nat Immunol 2002; 3: 1061– 1068. [DOI] [PubMed] [Google Scholar]
- 63. Goulder PJ, Bunce M, Krausa P et al. Novel, cross-restricted, conserved, and immunodominant cytotoxic T lymphocyte epitopes in slow progressors in HIV type 1 infection. AIDS Res Hum Retroviruses 1996; 12: 1691– 1698. [DOI] [PubMed] [Google Scholar]
- 64. Altfeld M, Addo MM, Rosenberg ES et al. Influence of HLA-B57 on clinical presentation and viral control during acute HIV-1 infection. AIDS 2003; 17: 2581– 2591. [DOI] [PubMed] [Google Scholar]
- 65. Dyer WB, Ogg GS, Demoitie M-A et al. Strong human immunodeficiency virus (HIV)-specific cytotoxic T-lymphocyte activity in Sydney Blood Bank Cohort patients infected with nef-defective HIV type 1. J Virol 1999; 73: 436– 443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Eriksson S, Graf EH, Dahl V et al. Comparative analysis of measures of viral reservoirs in HIV-1 eradication studies. PLoS Pathog 2013; 9: e1003174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hatano H, Delwart EL, Norris PJ et al. Evidence for persistent low-level viremia in individuals who control human immunodeficiency virus in the absence of antiretroviral therapy. J Virol 2009; 83: 329– 335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Mendoza D, Johnson SA, Peterson BA et al. Comprehensive analysis of unique cases with extraordinary control over HIV replication. Blood 2012; 119: 4645– 4655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Hunt PW, Brenchley J, Sinclair E et al. Relationship between T cell activation and CD4+ T cell count in HIV-seropositive individuals with undetectable plasma HIV RNA levels in the absence of therapy. J Infect Dis 2008; 197: 126– 133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Lafeuillade A, Poggi C, Tamalet C et al. Effects of a combination of zidovudine, didanosine, and lamivudine on primary human immunodeficiency virus type 1 infection. J Infect Dis 1997; 175: 1051– 1055. [DOI] [PubMed] [Google Scholar]
- 71. Zaunders JJ, Cunningham PH, Kelleher AD et al. Potent antiretroviral therapy of primary human immunodeficiency virus type 1 (HIV-1) infection: partial normalization of T lymphocyte subsets and limited reduction of HIV-1 DNA despite clearance of plasma viremia. J Infect Dis 1999; 180: 320– 329. [DOI] [PubMed] [Google Scholar]
- 72. Kassutto S, Johnston MN, Rosenberg ES. Incomplete HIV type 1 antibody evolution and seroreversion in acutely infected individuals treated with early antiretroviral therapy. Clin Infect Dis 2005; 40: 868– 873. [DOI] [PubMed] [Google Scholar]
- 73. Killian MS, Norris PJ, Rawal BD et al. The effects of early antiretroviral therapy and its discontinuation on the HIV-specific antibody response. AIDS Res Hum Retroviruses 2006; 22: 640– 647. [DOI] [PubMed] [Google Scholar]
- 74. Henrich TJ, Hu Z, Li JZ et al. Long-term reduction in peripheral blood HIV type 1 reservoirs following reduced-intensity conditioning allogeneic stem cell transplantation. J Infect Dis 2013; 207: 1694– 1702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. McMichael AJ, Rowland-Jones SL. Cellular immune responses to HIV. Nature 2001; 410: 980– 987. [DOI] [PubMed] [Google Scholar]
- 76. Reynolds MR, Weiler AM, Weisgrau KL et al. Macaques vaccinated with live-attenuated SIV control replication of heterologous virus. J Exp Med 2008; 205: 2537– 2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Barker E, Mackewicz CE, Reyes-Teran G et al. Virological and immunological features of long-term human immunodeficiency virus-infected individuals who have remained asymptomatic compared with those who have progressed to acquired immunodeficiency syndrome. Blood 1998; 92: 3105– 3114. [PubMed] [Google Scholar]
- 78. Saez-Cirion A, Lacabaratz C, Lambotte O et al. HIV controllers exhibit potent CD8 T cell capacity to suppress HIV infection ex vivo and peculiar cytotoxic T lymphocyte activation phenotype. Proc Natl Acad Sci U S A 2007; 104: 6776– 6781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Wilkinson J, Zaunders JJ, Carr A et al. Characterization of the phenotypic and lymphokine profile associated with strong CD8+ anti-HIV-1 suppressor activity (CASA). Clin Exp Immunol 2002; 127: 145– 150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Quiros JC, Shupert WL, McNeil AC et al. Resistance to replication of human immunodeficiency virus challenge in SCID-Hu mice engrafted with peripheral blood mononuclear cells of nonprogressors is mediated by CD8(+) T cells and associated with a proliferative response to p24 antigen. J Virol 2000; 74: 2023– 2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Martinez-Picado J, Prado JG, Fry EE et al. Fitness cost of escape mutations in p24 Gag in association with control of human immunodeficiency virus type 1. J Virol 2006; 80: 3617– 3623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Forshey BM, Schwedler U, Sundquist WI, Aiken C. Formation of a human immunodeficiency virus type 1 core of optimal stability is crucial for viral replication. J Virol 2002; 76: 5667– 5677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Norris PJ, Moffett HF, Brander C et al. Fine specificity and cross-clade reactivity of HIV type 1 Gag-specific CD4+ T cells. AIDS Res Hum Retroviruses 2004; 20: 315– 325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Lichterfeld M, Kaufmann DE, Yu XG et al. Loss of HIV-1-specific CD8+ T cell proliferation after acute HIV-1 infection and restoration by vaccine-induced HIV-1-specific CD4+ T cells. J Exp Med 2004; 200: 701– 712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Demkowicz WE Jr, Littaua RA, Wang J, Ennis FA. Human cytotoxic T-cell memory: long-lived responses to vaccinia virus. J Virol 1996; 70: 2627– 2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Altfeld M, Allen T. Hitting HIV where it hurts: an alternative approach to HIV vaccine design. Trends Immunol 2006; 27: 504– 510. [DOI] [PubMed] [Google Scholar]
- 87. Wang W, Zhou J, Halambage UD et al. Inhibition of HIV-1 maturation via small-molecule targeting of the amino-terminal domain in the viral capsid protein. J Virol 2017; 91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Huang SH, Ren Y, Thomas AS et al. Latent HIV reservoirs exhibit inherent resistance to elimination by CD8+ T cells. J Clin Invest 2018; 128: 876– 889. [DOI] [PMC free article] [PubMed] [Google Scholar]