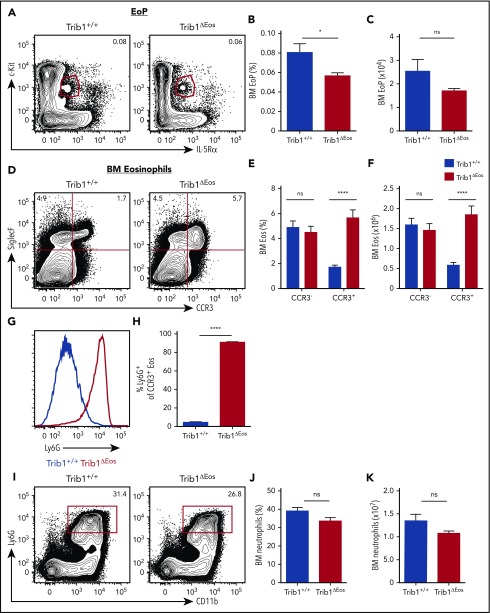
Figure 4.
Eosinophil-specific deletion of Trib1 results in Ly6G+ eosinophil development with improved preservation of EoPs. (A) Representative plots of BM EoP, gated on live, lineage−CD34+ cells from Trib1+/+ and Trib1ΔEos mice. Frequency of live cells (B) and absolute numbers (C) of BM EoP from Trib1+/+ and Trib1ΔEos mice; n = 6 mice per group, pooled from 2 experiments. (D) Representative plots of BM eosinophils from Trib1+/+ and Trib1ΔEos mice gated on live, CD11b+ cells. Frequency of live cells (E) and absolute numbers (F) of BM eosinophils by CCR3 expression. (G) Representative histogram of Ly6G expression, gated on live, CD11b+SiglecF+CCR3+, Trib1+/+ (blue), Trib1ΔHSC (red). (H) Frequency of SiglecF+CCR3+ cells expressing Ly6G; n = 11 mice per group, pooled from 4 experiments. (I) Representative plots of BM neutrophils from Trib1+/+ and Trib1ΔEos mice, gated on live, SiglecF− cells. (J) Frequency of live cells and (K) absolute numbers of BM neutrophils; n = 11 mice per group, pooled from 4 experiments. *P = .0289; ****P < .0001; unpaired Student t test. Frequencies and error bars are mean ± SEM of live cells.