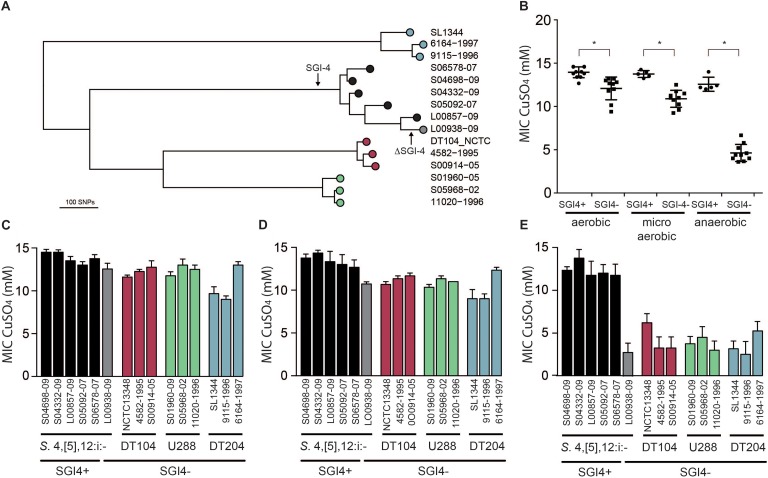
FIGURE 4.
Phylogenetic relationship and Cu sulfate MIC of representative strains of monophasic S. Typhimurium ST34 and S. Typhimurium. (A) A mid-point rooted phylogenetic tree constructed using sequence variation in the core genome with reference to S. Typhimurium strain SL1344 whole genome sequence (accession FQ312003). Leaves of the tree corresponding to representative strains are labeled as filled circles color coded for SGI-4+ monophasic S. Typhimurium ST34 (black) and SGI-4- monophasic S. Typhimurium ST34 (gray), and S. Typhimurium DT104 complex (red), U288 complex (green), and DT204 complex (blue) are shown. (B) Mean MIC for Cu sulfate of SGI-4+ (filled circles) and SGI-4- (filled squares) monophasic S. Typhimurium ST34 and S. Typhimurium strains during aerobic, microaerobic and anaerobic culture. Bars indicate the mean MIC for Cu sulfate for each strain ± standard deviation in aerobic (C), microaerobic (D), and anaerobic (E) atmosphere. Bar colors match the tree leaf circles in (A). ∗Indicates that groups indicated by lines were significantly different (p < 0.05).