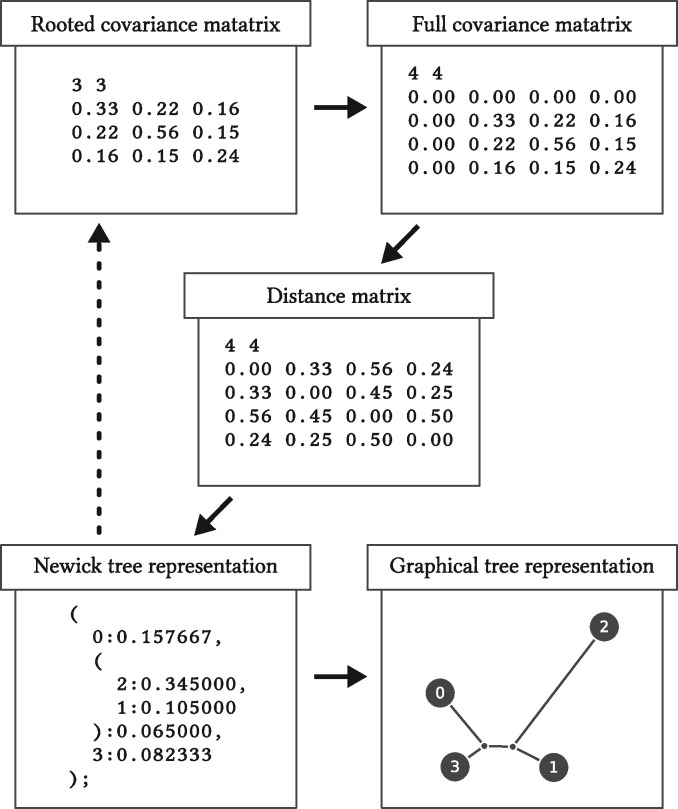
Fig. 1.
Phylogenetic tree construction pipeline. We estimate a rooted covariance matrix, where the root is arbitrarily chosen. We then recover the full covariance matrix, compute the distance matrix, and approximate the distance matrix as a tree structure using the NJ algorithm. Optionally, by supplying the estimated tree into the covariance inference process, we refine the estimated covariances to be most tree-compatible. Finally, we render the Newick tree in SVG format. For better control of the graphics, we recommend using our web service: http://www.jade-cheng.com/graphs/