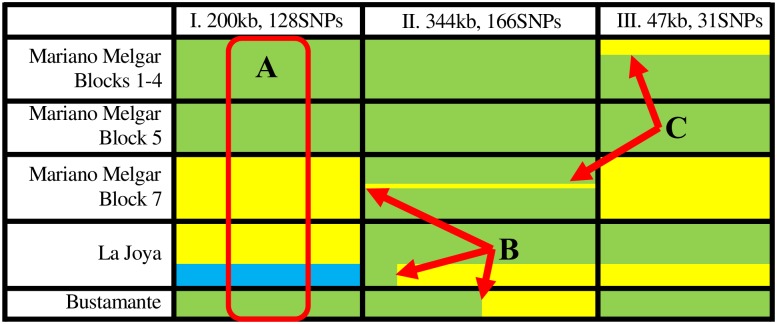
Fig 3. The distribution of polymorphic markers within and among individuals provides clear evidence of meiotic recombination.
This representative subset of the data portrays the distributions of 325 polymorphic markers throughout three regions of the genome in 54 isolates (grouped by geographic location: Mariano Melgar Blocks 1–4 (N = 14), Block 5 (N = 11), Block 7 (N = 14), La Joya (N = 14), and Bustamante (N = 1)) that indicate the presence of sexual reproduction as A) the independent assortment of chromosomes among individuals in a population; B) meiotic recombination events that reassort polymorphic markers along a chromosome; and C) evidence of very recent reassortment of polymorphic markers via sexual reproduction. Markers that are polymorphic in the population can be heterozygous (green) or homozygous in each individual (yellow representing a site that is homozygous for one base, blue representing individuals homozygous for the alternative base). For clarity, only polymorphic sites with a minor allele frequency >16% in the population are shown here. (A) Independent assortment of chromosomes and subsequent fusion of gametes has resulted in a region (I) that is homozygous in some individuals and heterozygous in others across a 200kb region containing 128 polymorphic markers on an 829kb contig. In this region, individuals can be homozygous for either set of linked polymorphic markers. Interestingly, genotypes are geographically clustered, indicating identity by descent. Runs of identical color indicate identical sequence. (B) Meiotic recombination has resulted in a region (II) of a 357kb contig that partially retain the linkage patterns of both parental chromosomes while the rest retains the linkage pattern of only one parental chromosome, showing how crossing over can affect parts of chromosomes. In this region, there is evidence of at least three independent meiotic recombination events. (C) Sexual reproduction after a recent colonization of a city block has resulted in the genetic divergence of closely related samples over a run of polymorphic markers. For example, a meiotic recombination event is apparent in region II in one sampled individual on Block 7. The remaining contigs (S1 Fig) contain near-complete sequence similarity among all clones isolated from Block 7 suggesting that all individuals on this block share a recent common ancestor and the meiotic event occurring in region II occurred after the Block 7 was colonized. Similarly, a meiotic recombination event in a region (III) of a 130kb contig occurred in one lineage inhabiting Blocks 1–3 in Mariano Melgar. The regions shown here contain only some of the 474 meiotic recombination events distributed across 151 contigs, with each detected meiotic recombination event spanning from 10kb to nearly 500kb. The relative positions of each contig within the genome are unknown. For data spanning the whole genome, see S1 Fig.