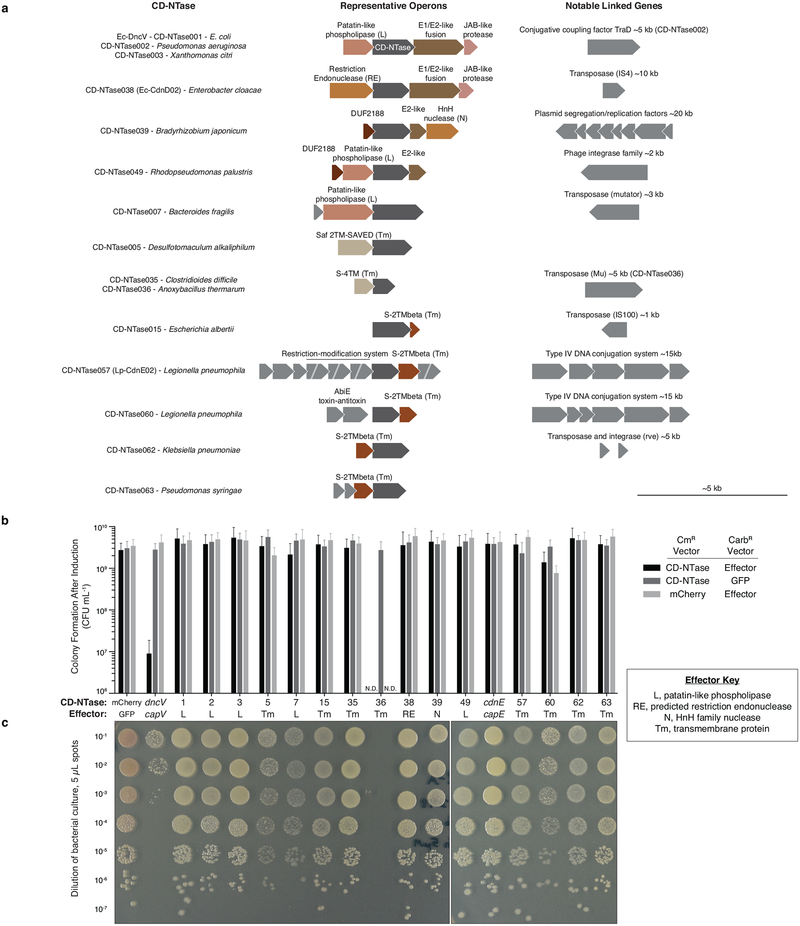
Extended Data Figure 8 |. CD-NTases are encoded in conserved, poorly understood operons on mobile genetic elements.
a-, Operon structure for CD-NTases selected for in-depth characterization showing the conserved protein domains in CD-NTase adjacent genes (see Fig. 4a–c). Conserved operons were first identified by Burroughs et al. and operons are vertically organized by similarity to one another22. Where found, linked genes demonstrating CD-NTases are encoded on mobile genetic elements are indicated.
b, CD-NTases and their adjacently encoded “effector” proteins were coexpressed in E. coli and bacterial colony formation was quantified by spot dilution analysis. CD-NTases were inducibly expressed from a chloramphenicol resistant (CmR) vector and effectors were inducibly expressed from a carbenicillin resistant (CarbR) vector. Bacteria harboring cognate CD-NTase / effector plasmids or control plasmids were plated on medium containing both inducers and incubated for 24 h at 37 °C. Data were not determined (N.D.) for CD-NTase036 because the effector was toxic to E. coli under non-inducing conditions. Data are the mean ± SEM of 3 independent experiments.
c, Spot dilution analysis of bacteria harboring the cognate CD-NTase-effector pair indicated, as quantified in b. The CD-NTase036-effector pair was not analyzed in this assay. Colony morphology indicates a potential interaction for some combinations, however, it is unclear how specific or significant this may be. Data are representative of 3 independent experiments.