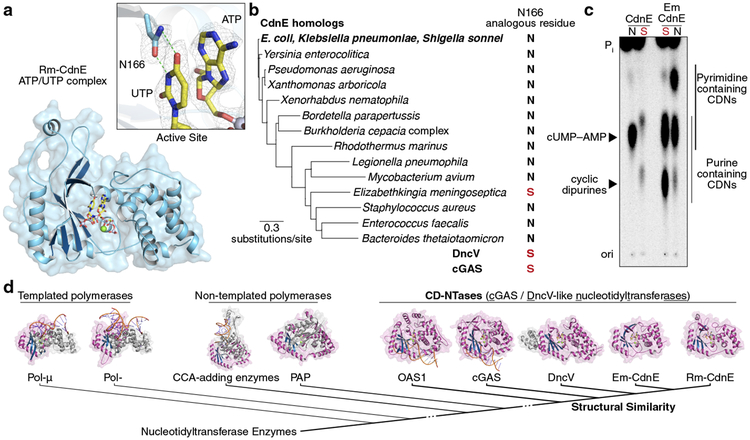
Figure 2 |. Conserved active site residues dictate CD-NTase specificity.
a, Crystal structure of Rm-CdnE in complex with nonhydrolyzable ATP and UTP analogs and zoom-in inset of key N166–uridine contacts controlling pyrimidine specificity. Green dotted lines indicate hydrogen bonding and 2Fo−Fc electron density map is contoured at 1 σ.
b, Phylogram of CdnE sequence homologs and their N166 analogous residue determined by sequence alignment (Extended Data Fig. 4a). Red “S’s” highlight cGAS / DncV-like serine residues.
c, CdnE homolog and mutant reactions analyzed by TLC as in Fig. 1b. “N” vs red “S” indicate asparagine or cGAS / DncV-like serine at the N166 analogous position in the tested allele. Side-chains are numbered according to Rm-CdnE sequence. Data are representative of 3 independent experiments. For detailed deconvolution and purine vs pyrimidine migration pattern analysis see Extended Data Fig. 3 and 4.
d, Structure-based comparisons define that Rm-CdnE and Em-CdnE are cGAS / DncV-like nucleotidyltransferase (CD-NTases) with a similar architecture to DncV (4TY0), cGAS (6CTA), and OAS1 (4RWO). CD-NTases are more distantly related to Pol-β-like NTases: Pol-μ (4YD1), Pol-β (4KLQ), CCA-adding enzyme (4X4T), and Poly(A) Polymerase gamma (PAP, 4LT6). The NTase core domain for each enzyme is clustered according to Z-score and colored in magenta/blue.