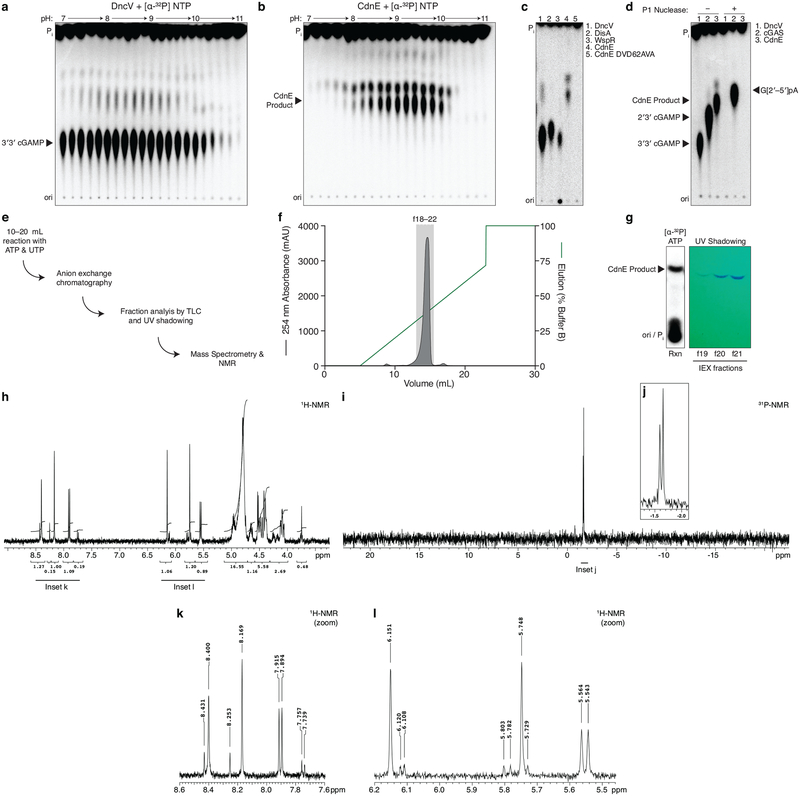
Extended Data Figure 1 |. Detailed characterization of CdnE, a cUMP–AMP synthase.
a and b, Titration of reaction buffer pH in steps of 0.2 pH units. Recombinant DncV or CdnE was incubated with α−32P radiolabeled ATP, CTP, GTP, and UTP at varying pH and the reactions were treated with alkaline phosphatase and visualized by PEI-cellulose TLC. CdnE activity is optimal at pH ~9.4, and this reaction condition was used in further experiments. Data are representative of 2 independent experiments.
c, PEI-cellulose TLC of products after incubation of indicated enzyme, wild-type CdnE, or active site mutant CdnE with α−32P radiolabeled ATP, CTP, GTP, and UTP as in Fig. 1b. Mutations that ablate the CdnE Mg2+-coordinating, active-site residues eliminate all detectable activity. Data are representative of 3 independent experiments.
d, Nuclease P1 sensitivity of CDN products. The endonuclease P1 specifically cleaves 3′–5′, canonical phosphodiester bonds. DncV and CdnE products are completely digested in the presence of P1 and alkaline phosphatase, whereas only one bond of 2′3′ cGAMP is susceptible to digestion, producing the linear G[2′–5′]pA product. Data are representative of 3 independent experiments.
e, Workflow of nucleotide production for MS and NMR analysis.
f, Anion exchange chromatography of a CdnE reaction with ATP and UTP, eluted with a gradient of Buffer B (2 M ammonium acetate) by FPLC. Individual fractions were concentrated prior to pooling for further analysis.
g, Anion exchange chromatography (IEX) fractions from f were separated by silica TLC, visualized by UV shadowing, and compared to a radiolabeled reaction to confirm the appropriate A254 peak. Fractions were pooled and concentrated prior to MS and NMR analysis.
h, k, and l, 3′3′ cyclic uridine monophosphate–adenosine monophosphate proton NMR spectra and associated zoomed in datasets. 1H NMR (400 MHz): δΗ 8.40 (s, 1H), 8.17 (s, 1H), 7.90 (d, J = 8.2 Hz, 1H), 6.15 (s, 1H), 5.75 (s, 1H), 5.55 (d, J = 8.2 Hz, 1H), 5.00–4.90 (m, 2H), 4.80 (d, J = 4.5 Hz, 1H) 4.70–4.61 (m, 1H), 4.55–4.38 (m, 4H), 4.17–4.02 (m, 2H)
i and j, 3′3′ cyclic uridine monophosphate–adenosine monophosphate phosphate NMR spectra and associated zoomed in dataset. 31P{1H} NMR (162 MHz): δP −1.59 (s, 1P), −1.65 (s, 1P).