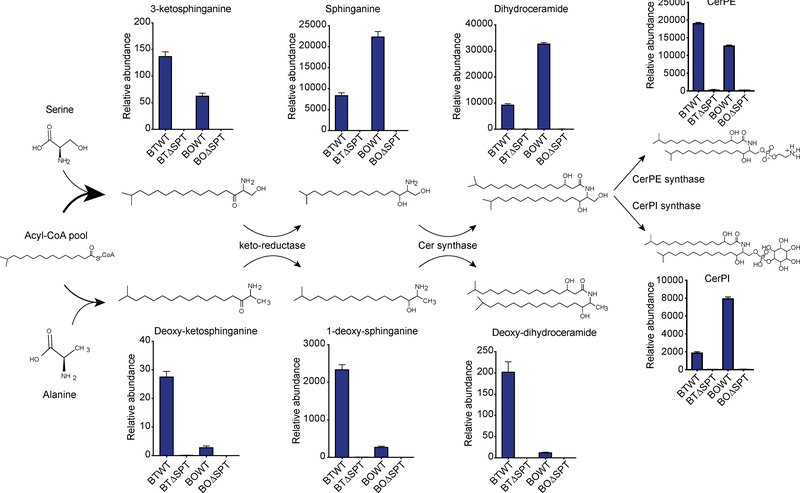
Figure 6: Proposed sphingolipid biosynthesis pathways for the most abundant sphingolipid metabolite features in Bacteroides.
Sphingolipid structures and features enriched in WT Bacteroides as predicted from LC-MS data. Bars represent relative abundances of each lipid in B. thetaiotaomicron and B. ovatus and losses in the corresponding ΔSPT strains. Annotation of SPT-dependent metabolite features reveals SPT can utilize serine (upper panels) or alanine (lower panels) to synthesize complex sphingolipids. From left to right, Spt is required for synthesis of 3-ketosphinganine or deoxy-ketosphinganine, sphinganine or 1-deoxy-sphinganine, dihydroceramide or deoxy-dihydroceramide, and ceramide phosphoethanolamine (CerPE) or ceramide phosphoinositol (CerPI). Chemical structure representations are based on an acyl chain backbone length of 17 carbons (C17) for sphingolipids and 18 carbons (C18) for deoxysphingolipids. See also Figures S5 and S6 and Data Files S3 and S5.