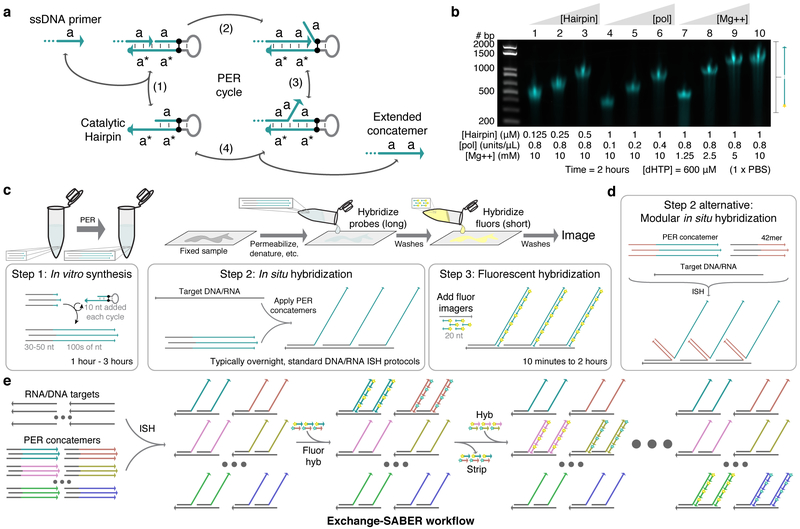
Figure 1: SABER-FISH design and workflow.
a, Primer exchange reaction (PER) cycle.30 A primer with domain a on its 3’ end binds (reversibly) to a catalytic hairpin (step 1) and gets extended with a new a domain by a strand displacing polymerase (step 2). Competitive branch migration (step 3)32 displaces the extended primer, which can then dissociate (step 4). Primers undergo the cycle repeatedly to form long repetitive concatemeric sequences. b, Length programmability of PER concatemers. Hairpin concentration, polymerase concentration, magnesium concentration, and incubation time can be adjusted to control PER kinetics and therefore concatemer length. Time and dHTP (dATP, dTTP, and dCTP) concentration were fixed at 2 hours and 600 μM, respectively. See also Supplementary Fig. 1d. c, SABER workflow. First, large batches of probe sequences with primers on their 3’ ends can be concatemerized using PER. These extensions are then hybridized to DNA and RNA targets in fixed samples following standard in situ hybridization (ISH) protocols. This is followed by a short step that hybridizes fluor- conjugated oligos (imagers) to complementary concatemers. d, An alternative method uses 42mer ‘bridge’ sequences to couple probes to concatemer sequences. This method allows the same 42mer barcodes with PER concatemers to be re-used for different targets. e, Exchange-SABER workflow. Optionally, fluorescent signal can be rapidly stripped from concatemers without disrupting probe binding, allowing exchange of imagers8,9,10 and reuse of the same fluorescence channels through multiple rounds of imaging.