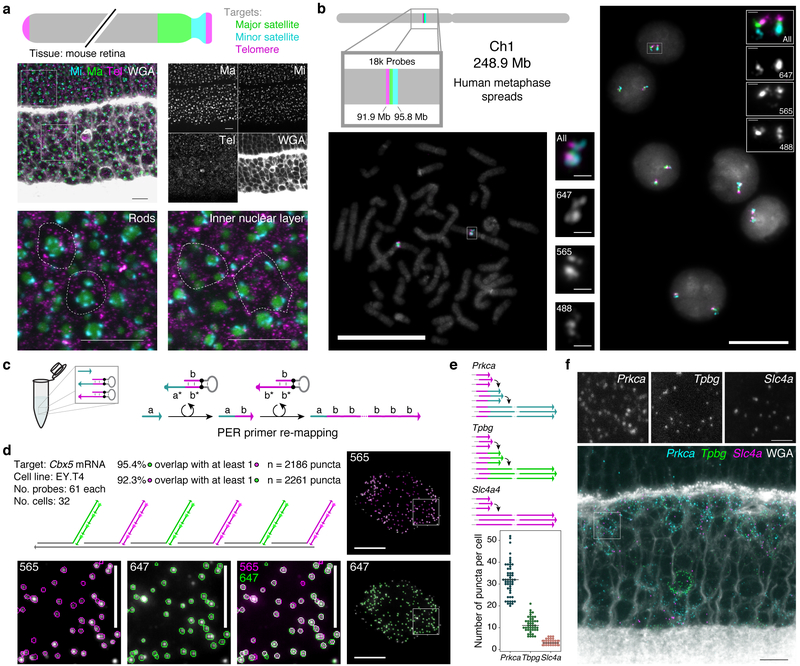
Figure 4: SABER enables spectrally multiplexed imaging.
a, Multiplexed SABER-FISH in mouse retina. Mouse major satellite, minor satellite, and telomere chromosomal regions were detected with orthogonal SABER concatemers. Magnified views show distinct organization of these chromosome regions in rods compared to inner nuclear layer cells. Dashed outlines indicate approximate cell boundaries based on WGA staining. Scale bars: 10 μm. b, Multiplexed SABER- FISH on metaphase spreads. Three adjacent positions on human chromosome 1 were visualized using a bridge strategy (Fig. 1d) in metaphase spreads and interphase cells. Scale bars: 20 μm. c, Primer re-mapping with PER. Primers (e.g. with domain a) can be concatemerized with a different repetitive sequence (e.g. primer domain b) using a stepwise PER hairpin (e.g. that appends b to a)30 and a standard repetitive hairpin. d, Single molecule co-localization. Primer re-mapping was used to map two halves of the Cbx5 probe pool (used in Fig. 2) to distinct concatemer sequences, and two-color co-localization was visualized (see also Supplementary Fig. 6b) and quantified. In total, 92.3% of identified puncta in the 565 channel ovelapped with puncta in 647, and 95.4% of identified puncta in 647 overlapped with puncta in the 565 channel. Scale bars: 10 μm (cells), 5 μm (panels). n(puncta): n(565)=2,261; n(647)=2,186. e, Primer re-mapping and 3-color visualization in retina tissue. The Tpbg and Prkca probe sets from Fig. 3 were re-mapped to two new primers to enable simultaneous visualization and quantification of Prkca, Tpbg, and Slc4a transcripts. Median lines shown on distributions. Representative images are depicted in panel f,. n(cells): n(Slc4a)=38; n(Tpbg)=45; n(Prkca)=52. Scale bars: 2.5 μm, 10 μm (overlay).