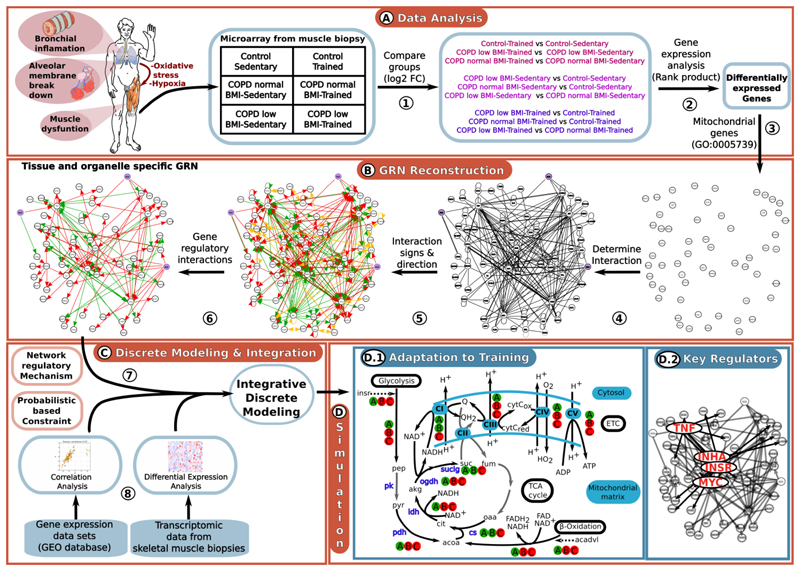
Fig. 1. Overall process of GRN reconstruction and its integration with probabilistic-based constraints into a discrete-model driven analysis.
Fig. 1A, step 1: Comparison of the trancriptomic data of the six study states Fig. 1A, step 2: Identify differentially expressed genes between conditions Fig. 1A, step 3: Since both energy metabolism and ROS production occur mainly in the mitochondria, the GRN reconstruction was focused on differentially expressed genes associated with mitochondrial processes using GO and Human Proteins Atlas databases. Fig. 1B, step 4: Automated interaction network reconstruction generated by IPA software (Krämer et al., 2014), DroID (Murali et al., 2011) and BioXM (Maier et al., 2011) databases based on the list of differentially expressed genes. This network includes gene regulatory and protein-protein interactions. Fig. 1B, step 5: Manual curation of the GRN reconstruction using a bibliomic data sources (Supplementary material 1.2) to identify and correct incomplete or erroneous annotations and identify the direction and sign of interactions. Fig. 1B, step 6: Since our analysis is based on transcriptomic data, we focused on gene regulatory interactions only. The nodes in the networks represent genes that are either differentialy expressed between groups or were included to construct a single GRN; violet nodes represent ROS and edges represent interactions between nodes, with green and red arrows indicating negative and positive interactions, respectively. Interactions with unknown direction and/or sign are in yellow. Fig. 1C step 7: Mathematical formalization of the reconstructed GRN based on R. Thomas formalism (Thomas and Kaufman, 2001). This approach describes mechanistically the interactions between differentially expressed mitochondrial-associated genes. Fig. 1C step 8: Impose constraints based on publicly available muscle-related gene expression data-sets Fig D.1: Applying the discrete model which integrates the gene-regulatory mechanisms (step 7) and the constraints (step 8) to determine the mechanisms associated with abnormal adaptation to training in COPD patients. Graphical representation of the predicted metabolic adaptation to endurance training program in healthy people (A), COPD patients with normal BMI (B) and COPD patients with low BMI (C). The figure represents a scheme of glycolysis, β-oxidation, TCA cycle and electric transport chain (ETC). Continuous arrows represent the metabolic reactions, while dashed arrows represent the activation of glycolysis and β-oxidation by insr and acadvl, respectively. Metabolic reactions/pathways associated with genes predicted to be up-regulated (down-regulated) after the training program are colored in green (red). White arrows represent metabolic reactions not directly associated with any gene in the reconstructed GRN. Gene abbreviations: insr: insulin receptor factor, acadvl: acyl-CoA dehydrogenase, very long chain. Metabolic reaction abbreviations: pk: pyruvare kinase, pdh: pyruvate dehydrogenase, cs: citrase synthase, idh: isocitrate dehydrogenase, ogdh: oxoglutarate dehydrogenase, suclg: succinate-CoA ligase, CI: NADH dehydrogenase, CII: succinate dehydrogenase, CIII: cytochrome c reductase, CIV: cytochrome c oxidase, CV: ATP synthase; the metabolites and cofactors represented in this figure are: pep: phosphoenol pyruvate, pyr: pyruvate, acoa: acetyl CoA, cit: citrate, akg: alpha-ketoglutarate, suc: succinate, fum: fumarate, oaa, oxalacetate, H+: proton, NAD+: nicotinamide adenine dinucleotide oxidized, NADH: nicotinamide adenine dinucleotide reduced, FAD: flavin adenine dinucleotide oxidized, FADH2: flavin adenine dinucleotide reduced, ATP: adenosine triphosphate, ADP: adenosine diphosphate, Q: ubiquinol, QH2: ubiquinone, cytCox: cytochrome c oxidized, cytCred: cytochrome c reduced. Fig D.2: Prediction of key regulators underlying the abnormal adaptation to training in COPD.