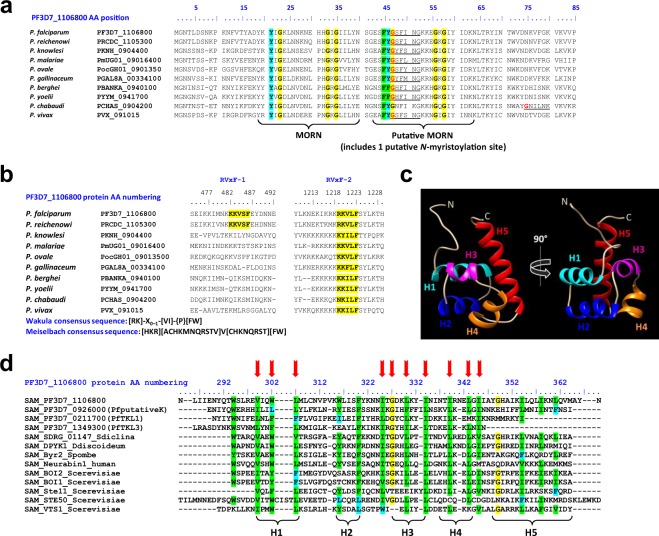
Figure 2.
In silico annotation of PfpTKL interaction motifs and domains. (a) Plasmodium PfpTKL homologs share common N-terminal features beside the MORN motif(s). The alignment was created with MAFFT89 and residues 100% conserved over all of the sequences were shaded using BioEdit v7.2.5116. The putative N-myristoylation site is shown in red. (b) Plasmodium PfpTKL homologs possess one or two RVxF motifs. The alignment was created with MAFFT89. RVxF motif consensus sequences used in silico to detect RVxF1 and RVxF2 are displayed below the alignment86,87. (c) The PfpTKL SAM undergoes classic folding. Tertiary structure PfpTKL SAM predicted with Phyre281 (75 residues (97%) modeled at >90% accuracy). Structure annotation and shading were achieved with Chimera v1.10.1117. (d) Plasmodium SAMs showing classic composition compared with SAM references. Sequence alignment created with MAFFT89 between the four P. falciparum SAMs and reference SAMs (Sdiclina = Saprolegnia diclina, Scerevisiae = Saccharomyces cerevisiae, Spombe = Schizosaccharomyces pombe, Ddiscoideum = Dictyostelium discoideum). Conserved residues were shaded using BioEdit v7.2.5116. Arrows indicate residues conserved in SAMs. Below the alignment are delineated the five α-helices (H1-H5) of the SAM core118.