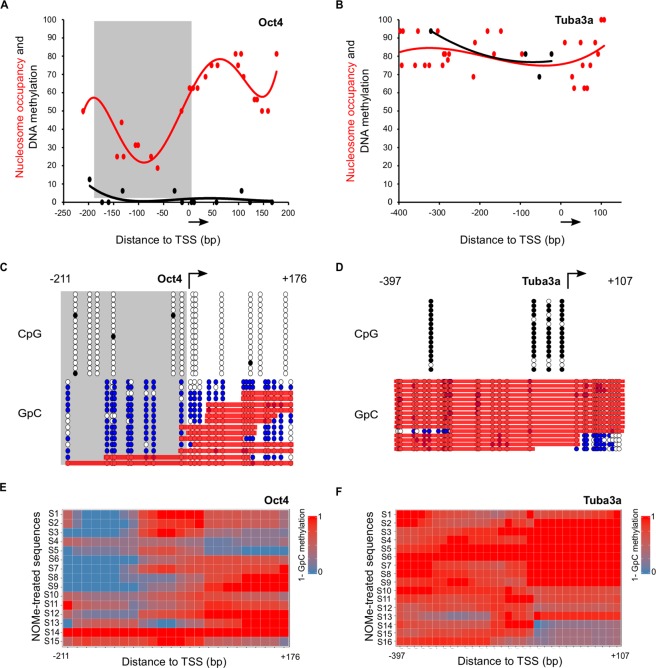
Figure 2.
Analysis of Oct4 and Tuba3a gene promoters using NOMe-PCR followed by NOMePlot in mESCs. (A,B) Scatter plots with trendline showing population-averaged nucleosome occupancy (red) and DNA methylation (black) relative to the TSS of an active gene (Oct4) (A) and a DNA methylated silent gene (Tuba3a) (B). Nucleosome occupancy is calculated as the percentage of unmethylated GpC sites. DNA methylation is plotted as the percentage of methylated CpG dinucleotides. Arrow indicates the direction of transcription. (C,D) Lollipop diagram showing DNA methylation and nucleosome occupancy at the single molecule for Oct4 (C) and Tuba3a (D). Top panels show cytosine methylation at CpGs (white for unmethylated, black for methylated) and bottom panels show methylation patterns at GpC dinucleotides (white for unmethylated, blue for methylated). Each line represents one molecule and each row correspond to a genomic position around the TSS (arrow). Red bars highlight GpC unmethylated regions long enough to accommodate a nucleosome. NDRs are marked with a grey box. (E,F) Heatmap of nucleosome occupancy for Oct4 (E) and Tuba3a (F) upon clustering of fifteen and sixteen sequences respectively. Nucleosome occupancy was calculated for each position around the TSS (X axis) as the average value of unmethylated cytosines at GpCs found within 140 bp windows. Color code goes from red (100% occupancy, 1-GpG = 1) to blue (0% occupancy, 1-GpC = 0).