Figure 1.
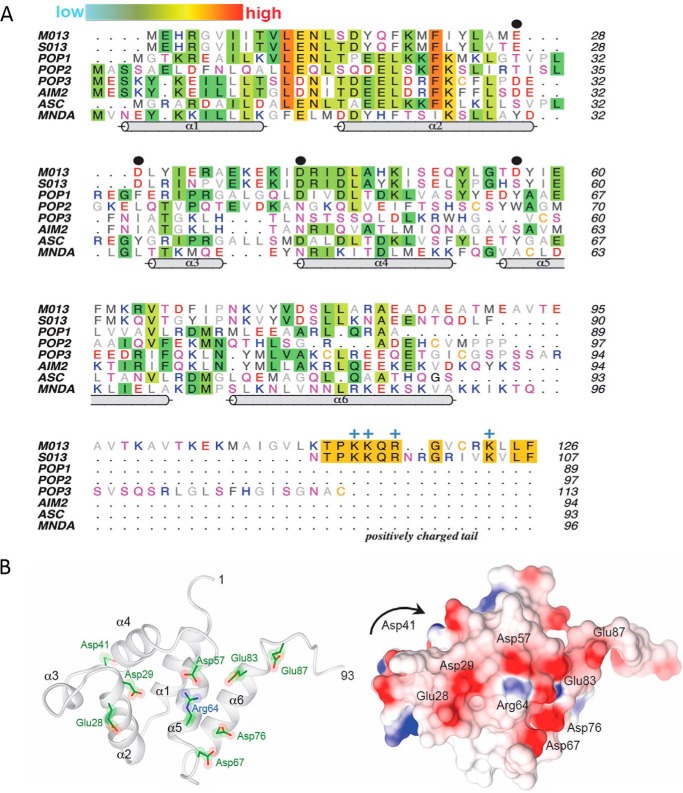
A, multiple sequence alignment of viral and cellular pyrin domains. Amino acids are colored by their aliphatic (gray) or hydrophilic properties (red, negative; blue, positive; magenta, polar), except where sequence conservation is observed (background shading). The relative extent of conservation is represented as a color gradient from cyan (low) to red (high). The PYD helices (cylinders) correspond to the structure of myeloid cell nuclear differentiation antigen (MNDA; Protein Data Bank code 2DBG). Black dots denote the key negatively charged residues (Glu-28, Asp-29, Asp-41, and Asp-57) that are discussed under “Results.” The positively charged tail of M013 and S013 is annotated along with the conserved Arg/Lys residues (blue +). B, homology modeling of the pyrin domain of M013. Left panel, ribbon model of the six α-helices (residues 1–93). Right panel, corresponding surface electrostatic representation of the same view, with red and blue indicating negative and positive charges, respectively. The view is looking direct at the type Ib face of the pyrin domain. Asp-41 resides at the end of α4 on the opposite face from the current view and is not visible on the electrostatic surface (denoted by the arrow).