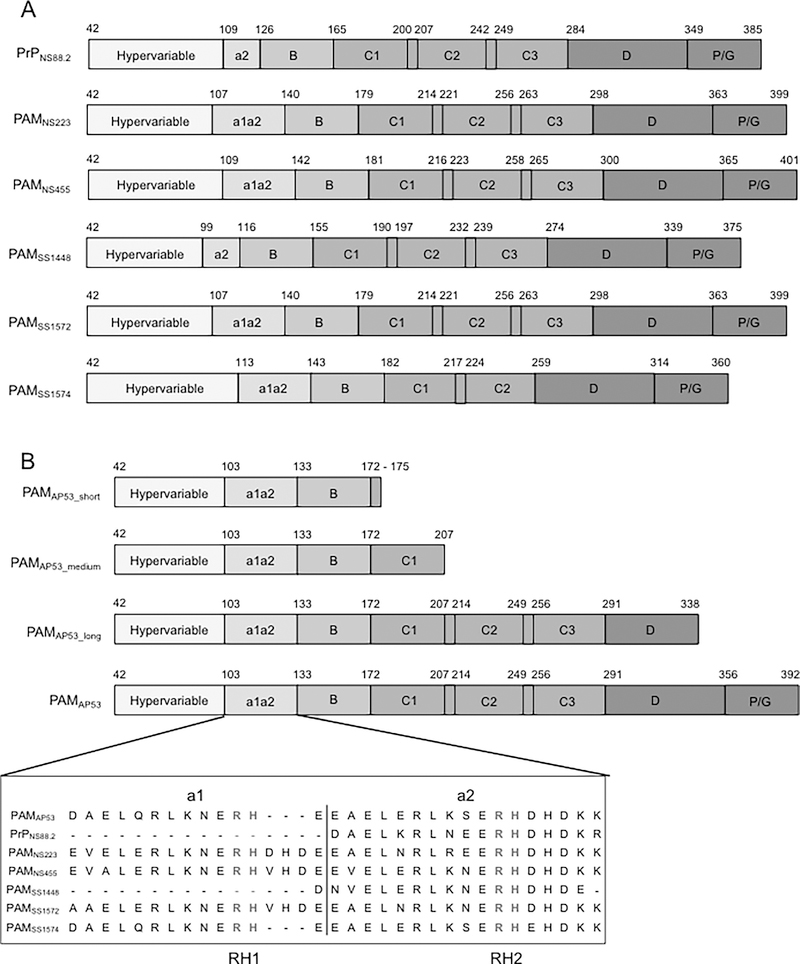
Fig. 1.
Schematics of full-length r-PAMs and truncated r-PAMAP53 peptides. All recombinant proteins start with the first residue (#42) immediately after the signal peptide. (A) Within all naturally occurring PAMs, homology progressively increases from their N- to C-termini. Domains align in the following order from the N-terminus: hypervariable region (HVR), A-domai n containing a1a2- or a2-repeats, B-domain, C-domain containing c1-c3 repeats, D-domain, and the Pro/Gly (P/G)-rich region. (B) Three truncated PAMAP53 proteins were prepared that terminated at different locations in the PAMAP53 sequence. The a1a2-repeats are zoomed in to manifest the variability of this domain among different r-PAMs. Residues in red refer to the sequence of a1a2-repeats, and residues in blue indicate the Arg-His (RH) motifs that specifically binding to hPg (Rios-Steiner et al., 2001; Sanderson-Smith et al., 2006b; Schenone et al., 2000; Wang et al., 2010a; Wang et al., 2010b). Dashes are applied to increase the homology of alignment when necessary. The schematic is drawn to relative scale, and numbers indicate the first and last residue of each domain.